Introducing the Shell
Overview
Teaching: 5 min
Exercises: 0 minQuestions
What is a command shell and why would I use one?
Objectives
Explain how the shell relates to the keyboard, the screen, the operating system, and users’ programs.
Explain when and why command-line interfaces should be used instead of graphical interfaces.
Background
Humans and computers commonly interact in many different ways, such as through a keyboard and mouse, touch screen interfaces, or using speech recognition systems. The most widely used way to interact with personal computers is called a graphical user interface (GUI). With a GUI, we give instructions by clicking a mouse and using menu-driven interactions.
While the visual aid of a GUI makes it intuitive to learn, this way of delivering instructions to a computer scales very poorly. Imagine the following task: for a literature search, you have to copy the third line of one thousand text files in one thousand different directories and paste it into a single file. Using a GUI, you would not only be clicking at your desk for several hours, but you could potentially also commit an error in the process of completing this repetitive task. This is where we take advantage of the Unix shell. The Unix shell is both a command-line interface (CLI) and a scripting language, allowing such repetitive tasks to be done automatically and fast. With the proper commands, the shell can repeat tasks with or without some modification as many times as we want. Using the shell, the task in the literature example can be accomplished in seconds.
The Shell
The shell is a program where users can type commands. With the shell, it’s possible to invoke complicated programs like climate modeling software or simple commands that create an empty directory with only one line of code. The most popular Unix shell is Bash (the Bourne Again SHell — so-called because it’s derived from a shell written by Stephen Bourne). Bash is the default shell on most modern implementations of Unix and in most packages that provide Unix-like tools for Windows.
Using the shell will take some effort and some time to learn. While a GUI presents you with choices to select, CLI choices are not automatically presented to you, so you must learn a few commands like new vocabulary in a language you’re studying. However, unlike a spoken language, a small number of “words” (i.e. commands) gets you a long way, and we’ll cover those essential few today.
The grammar of a shell allows you to combine existing tools into powerful pipelines and handle large volumes of data automatically. Sequences of commands can be written into a script, improving the reproducibility of workflows.
In addition, the command line is often the easiest way to interact with remote machines and supercomputers. Familiarity with the shell is near essential to run a variety of specialized tools and resources including high-performance computing systems. As clusters and cloud computing systems become more popular for scientific data crunching, being able to interact with the shell is becoming a necessary skill. We can build on the command-line skills covered here to tackle a wide range of scientific questions and computational challenges.
Let’s get started.
When the shell is first opened, you are presented with a prompt, indicating that the shell is waiting for input.
$
The shell typically uses $ as the prompt, but may use a different symbol.
In the examples for this lesson, we’ll show the prompt as $ .
Most importantly:
when typing commands, either from these lessons or from other sources,
do not type the prompt, only the commands that follow it.
Also note that after you type a command, you have to press the Enter key to execute it.
The prompt is followed by a text cursor, a character that indicates the position where your typing will appear. The cursor is usually a flashing or solid block, but it can also be an underscore or a pipe. You may have seen it in a text editor program, for example.
So let’s try our first command, ls which is short for listing.
This command will list the contents of the current directory:
$ ls
Desktop Downloads Movies Pictures
Documents Library Music Public
Command not found
If the shell can’t find a program whose name is the command you typed, it will print an error message such as:
$ ksks: command not foundThis might happen if the command was mis-typed or if the program corresponding to that command is not installed.
Nelle’s Pipeline: A Typical Problem
Nelle Nemo, a marine biologist,
has just returned from a six-month survey of the
North Pacific Gyre,
where she has been sampling gelatinous marine life in the
Great Pacific Garbage Patch.
She has 1520 samples that she’s run through an assay machine to measure the relative abundance
of 300 proteins.
She needs to run these 1520 files through an imaginary program called goostats.sh she inherited.
On top of this huge task, she has to write up results by the end of the month so her paper
can appear in a special issue of Aquatic Goo Letters.
The bad news is that if she has to run goostats.sh by hand using a GUI,
she’ll have to select and open a file 1520 times.
If goostats.sh takes 30 seconds to run each file, the whole process will take more than 12 hours
of Nelle’s attention.
With the shell, Nelle can instead assign her computer this mundane task while she focuses
her attention on writing her paper.
The next few lessons will explore the ways Nelle can achieve this.
More specifically,
they explain how she can use a command shell to run the goostats.sh program,
using loops to automate the repetitive steps of entering file names,
so that her computer can work while she writes her paper.
As a bonus, once she has put a processing pipeline together, she will be able to use it again whenever she collects more data.
In order to achieve her task, Nelle needs to know how to:
- navigate to a file/directory
- create a file/directory
- check the length of a file
- chain commands together
- retrieve a set of files
- iterate over files
- run a shell script containing her pipeline
Key Points
A shell is a program whose primary purpose is to read commands and run other programs.
This lesson uses Bash, the default shell in many implementations of Unix.
Programs can be run in Bash by entering commands at the command-line prompt.
The shell’s main advantages are its high action-to-keystroke ratio, its support for automating repetitive tasks, and its capacity to access networked machines.
The shell’s main disadvantages are its primarily textual nature and how cryptic its commands and operation can be.
Navigating Files and Directories
Overview
Teaching: 30 min
Exercises: 10 minQuestions
How can I move around on my computer?
How can I see what files and directories I have?
How can I specify the location of a file or directory on my computer?
Objectives
Explain the similarities and differences between a file and a directory.
Translate an absolute path into a relative path and vice versa.
Construct absolute and relative paths that identify specific files and directories.
Use options and arguments to change the behaviour of a shell command.
Demonstrate the use of tab completion and explain its advantages.
The part of the operating system responsible for managing files and directories is called the file system. It organizes our data into files, which hold information, and directories (also called ‘folders’), which hold files or other directories.
Several commands are frequently used to create, inspect, rename, and delete files and directories. To start exploring them, we’ll go to our open shell window.
First, let’s find out where we are by running a command called pwd
(which stands for ‘print working directory’). Directories are like places — at any time
while we are using the shell, we are in exactly one place called
our current working directory. Commands mostly read and write files in the
current working directory, i.e. ‘here’, so knowing where you are before running
a command is important. pwd shows you where you are:
$ pwd
/Users/nelle
Here,
the computer’s response is /Users/nelle,
which is Nelle’s home directory:
Home Directory Variation
The home directory path will look different on different operating systems. On Linux, it may look like
/home/nelle, and on Windows, it will be similar toC:\Documents and Settings\nelleorC:\Users\nelle. (Note that it may look slightly different for different versions of Windows.) In future examples, we’ve used Mac output as the default - Linux and Windows output may differ slightly but should be generally similar.We will also assume that your
pwdcommand returns your user’s home directory. Ifpwdreturns something different, you may need to navigate there usingcdor some commands in this lesson will not work as written. See Exploring Other Directories for more details on thecdcommand.
To understand what a ‘home directory’ is, let’s have a look at how the file system as a whole is organized. For the sake of this example, we’ll be illustrating the filesystem on our scientist Nelle’s computer. After this illustration, you’ll be learning commands to explore your own filesystem, which will be constructed in a similar way, but not be exactly identical.
On Nelle’s computer, the filesystem looks like this:
At the top is the root directory
that holds everything else.
We refer to it using a slash character, /, on its own;
this character is the leading slash in /Users/nelle.
Inside that directory are several other directories:
bin (which is where some built-in programs are stored),
data (for miscellaneous data files),
Users (where users’ personal directories are located),
tmp (for temporary files that don’t need to be stored long-term),
and so on.
We know that our current working directory /Users/nelle is stored inside /Users
because /Users is the first part of its name.
Similarly,
we know that /Users is stored inside the root directory /
because its name begins with /.
Slashes
Notice that there are two meanings for the
/character. When it appears at the front of a file or directory name, it refers to the root directory. When it appears inside a path, it’s just a separator.
Underneath /Users,
we find one directory for each user with an account on Nelle’s machine,
her colleagues imhotep and larry.
The user imhotep’s files are stored in /Users/imhotep,
user larry’s in /Users/larry,
and Nelle’s in /Users/nelle. Because Nelle is the user in our
examples here, therefore we get /Users/nelle as our home directory.
Typically, when you open a new command prompt, you will be in
your home directory to start.
Now let’s learn the command that will let us see the contents of our
own filesystem. We can see what’s in our home directory by running ls:
$ ls
Applications Documents Library Music Public
Desktop Downloads Movies Pictures
(Again, your results may be slightly different depending on your operating system and how you have customized your filesystem.)
ls prints the names of the files and directories in the current directory.
We can make its output more comprehensible by using the -F option
which tells ls to classify the output
by adding a marker to file and directory names to indicate what they are:
- a trailing
/indicates that this is a directory @indicates a link*indicates an executable
Depending on your shell’s default settings, the shell might also use colors to indicate whether each entry is a file or directory.
$ ls -F
Applications/ Documents/ Library/ Music/ Public/
Desktop/ Downloads/ Movies/ Pictures/
Here, we can see that our home directory contains only sub-directories. Any names in our output that don’t have a classification symbol are plain old files.
Clearing your terminal
If your screen gets too cluttered, you can clear your terminal using the
clearcommand. You can still access previous commands using ↑ and ↓ to move line-by-line, or by scrolling in your terminal.
Getting help
ls has lots of other options. There are two common ways to find out how
to use a command and what options it accepts —
depending on your environment, you might find that only one of these ways works:
- We can pass a
--helpoption to the command (not available on macOS), such as:$ ls --help - We can read its manual with
man(not available in Git Bash), such as:$ man ls
We’ll describe both ways next.
The --help option
Most bash commands and programs that people have written to be
run from within bash, support a --help option that displays more
information on how to use the command or program.
$ ls --help
Usage: ls [OPTION]... [FILE]...
List information about the FILEs (the current directory by default).
Sort entries alphabetically if neither -cftuvSUX nor --sort is specified.
Mandatory arguments to long options are mandatory for short options, too.
-a, --all do not ignore entries starting with .
-A, --almost-all do not list implied . and ..
--author with -l, print the author of each file
-b, --escape print C-style escapes for nongraphic characters
--block-size=SIZE scale sizes by SIZE before printing them; e.g.,
'--block-size=M' prints sizes in units of
1,048,576 bytes; see SIZE format below
-B, --ignore-backups do not list implied entries ending with ~
-c with -lt: sort by, and show, ctime (time of last
modification of file status information);
with -l: show ctime and sort by name;
otherwise: sort by ctime, newest first
-C list entries by columns
--color[=WHEN] colorize the output; WHEN can be 'always' (default
if omitted), 'auto', or 'never'; more info below
-d, --directory list directories themselves, not their contents
-D, --dired generate output designed for Emacs' dired mode
-f do not sort, enable -aU, disable -ls --color
-F, --classify append indicator (one of */=>@|) to entries
... ... ...
Unsupported command-line options
If you try to use an option that is not supported,
lsand other commands will usually print an error message similar to:$ ls -jls: invalid option -- 'j' Try 'ls --help' for more information.
The man command
The other way to learn about ls is to type
$ man ls
This command will turn your terminal into a page with a description
of the ls command and its options.
To navigate through the man pages,
you may use ↑ and ↓ to move line-by-line,
or try B and Spacebar to skip up and down by a full page.
To search for a character or word in the man pages,
use / followed by the character or word you are searching for.
Sometimes a search will result in multiple hits.
If so, you can move between hits using N (for moving forward) and
Shift+N (for moving backward).
To quit the man pages, press Q.
Manual pages on the web
Of course, there is a third way to access help for commands: searching the internet via your web browser. When using internet search, including the phrase
unix man pagein your search query will help to find relevant results.GNU provides links to its manuals including the core GNU utilities, which covers many commands introduced within this lesson.
Exploring More
lsFlagsYou can also use two options at the same time. What does the command
lsdo when used with the-loption? What about if you use both the-land the-hoption?Some of its output is about properties that we do not cover in this lesson (such as file permissions and ownership), but the rest should be useful nevertheless.
Solution
The
-loption makeslsuse a long listing format, showing not only the file/directory names but also additional information, such as the file size and the time of its last modification. If you use both the-hoption and the-loption, this makes the file size ‘human readable’, i.e. displaying something like5.3Kinstead of5369.
Listing in Reverse Chronological Order
By default,
lslists the contents of a directory in alphabetical order by name. The commandls -tlists items by time of last change instead of alphabetically. The commandls -rlists the contents of a directory in reverse order. Which file is displayed last when you combine the-tand-roptions? Hint: You may need to use the-loption to see the last changed dates.Solution
The most recently changed file is listed last when using
-rt. This can be very useful for finding your most recent edits or checking to see if a new output file was written.
Exploring Other Directories
Not only can we use ls on the current working directory,
but we can use it to list the contents of a different directory.
Let’s take a look at our Desktop directory by running ls -F Desktop,
i.e.,
the command ls with the -F option and the argument Desktop.
The argument Desktop tells ls that
we want a listing of something other than our current working directory:
$ ls -F Desktop
shell-lesson-data/
Note that if a directory named Desktop does not exist in your current working directory,
this command will return an error. Typically, a Desktop directory exists in your
home directory, which we assume is the current working directory of your bash shell.
Your output should be a list of all the files and sub-directories in your
Desktop directory, including the shell-lesson-data directory you downloaded at
the setup for this lesson.
On many systems,
the command line Desktop directory is the same as your GUI Desktop.
Take a look at your Desktop to confirm that your output is accurate.
As you may now see, using a bash shell is strongly dependent on the idea that your files are organized in a hierarchical file system. Organizing things hierarchically in this way helps us keep track of our work: it’s possible to put hundreds of files in our home directory, just as it’s possible to pile hundreds of printed papers on our desk, but it’s a self-defeating strategy.
Now that we know the shell-lesson-data directory is located in our Desktop directory, we
can do two things.
First, we can look at its contents, using the same strategy as before, passing
a directory name to ls:
$ ls -F Desktop/shell-lesson-data
exercise-data/ north-pacific-gyre/
Second, we can actually change our location to a different directory, so we are no longer located in our home directory.
The command to change locations is cd followed by a
directory name to change our working directory.
cd stands for ‘change directory’,
which is a bit misleading:
the command doesn’t change the directory;
it changes the shell’s idea of what directory we are in.
The cd command is akin to double clicking a folder in a graphical interface to get into a folder.
Let’s say we want to move to the data directory we saw above. We can
use the following series of commands to get there:
$ cd Desktop
$ cd shell-lesson-data
$ cd exercise-data
These commands will move us from our home directory into our Desktop directory, then into
the shell-lesson-data directory, then into the exercise-data directory.
You will notice that cd doesn’t print anything. This is normal.
Many shell commands will not output anything to the screen when successfully executed.
But if we run pwd after it, we can see that we are now
in /Users/nelle/Desktop/shell-lesson-data/exercise-data.
If we run ls -F without arguments now,
it lists the contents of /Users/nelle/Desktop/shell-lesson-data/exercise-data,
because that’s where we now are:
$ pwd
/Users/nelle/Desktop/shell-lesson-data/exercise-data
$ ls -F
animal-counts/ creatures/ numbers.txt proteins/ writing/
We now know how to go down the directory tree (i.e. how to go into a subdirectory), but how do we go up (i.e. how do we leave a directory and go into its parent directory)? We might try the following:
$ cd shell-lesson-data
-bash: cd: shell-lesson-data: No such file or directory
But we get an error! Why is this?
With our methods so far,
cd can only see sub-directories inside your current directory. There are
different ways to see directories above your current location; we’ll start
with the simplest.
There is a shortcut in the shell to move up one directory level that looks like this:
$ cd ..
.. is a special directory name meaning
“the directory containing this one”,
or more succinctly,
the parent of the current directory.
Sure enough,
if we run pwd after running cd .., we’re back in /Users/nelle/Desktop/shell-lesson-data:
$ pwd
/Users/nelle/Desktop/shell-lesson-data
The special directory .. doesn’t usually show up when we run ls. If we want
to display it, we can add the -a option to ls -F:
$ ls -F -a
./ ../ exercise-data/ north-pacific-gyre/
-a stands for ‘show all’;
it forces ls to show us file and directory names that begin with .,
such as .. (which, if we’re in /Users/nelle, refers to the /Users directory).
As you can see,
it also displays another special directory that’s just called .,
which means ‘the current working directory’.
It may seem redundant to have a name for it,
but we’ll see some uses for it soon.
Note that in most command line tools, multiple options can be combined
with a single - and no spaces between the options: ls -F -a is
equivalent to ls -Fa.
Other Hidden Files
In addition to the hidden directories
..and., you may also see a file called.bash_profile. This file usually contains shell configuration settings. You may also see other files and directories beginning with.. These are usually files and directories that are used to configure different programs on your computer. The prefix.is used to prevent these configuration files from cluttering the terminal when a standardlscommand is used.
These three commands are the basic commands for navigating the filesystem on your computer:
pwd, ls, and cd. Let’s explore some variations on those commands. What happens
if you type cd on its own, without giving
a directory?
$ cd
How can you check what happened? pwd gives us the answer!
$ pwd
/Users/nelle
It turns out that cd without an argument will return you to your home directory,
which is great if you’ve got lost in your own filesystem.
Let’s try returning to the exercise-data directory from before. Last time, we used
three commands, but we can actually string together the list of directories
to move to exercise-data in one step:
$ cd Desktop/shell-lesson-data/exercise-data
Check that we’ve moved to the right place by running pwd and ls -F.
If we want to move up one level from the data directory, we could use cd ... But
there is another way to move to any directory, regardless of your
current location.
So far, when specifying directory names, or even a directory path (as above),
we have been using relative paths. When you use a relative path with a command
like ls or cd, it tries to find that location from where we are,
rather than from the root of the file system.
However, it is possible to specify the absolute path to a directory by
including its entire path from the root directory, which is indicated by a
leading slash. The leading / tells the computer to follow the path from
the root of the file system, so it always refers to exactly one directory,
no matter where we are when we run the command.
This allows us to move to our shell-lesson-data directory from anywhere on
the filesystem (including from inside exercise-data). To find the absolute path
we’re looking for, we can use pwd and then extract the piece we need
to move to shell-lesson-data.
$ pwd
/Users/nelle/Desktop/shell-lesson-data/exercise-data
$ cd /Users/nelle/Desktop/shell-lesson-data
Run pwd and ls -F to ensure that we’re in the directory we expect.
Two More Shortcuts
The shell interprets a tilde (
~) character at the start of a path to mean “the current user’s home directory”. For example, if Nelle’s home directory is/Users/nelle, then~/datais equivalent to/Users/nelle/data. This only works if it is the first character in the path:here/there/~/elsewhereis nothere/there/Users/nelle/elsewhere.Another shortcut is the
-(dash) character.cdwill translate-into the previous directory I was in, which is faster than having to remember, then type, the full path. This is a very efficient way of moving back and forth between two directories – i.e. if you executecd -twice, you end up back in the starting directory.The difference between
cd ..andcd -is that the former brings you up, while the latter brings you back.
Try it! First navigate to
~/Desktop/shell-lesson-data(you should already be there).$ cd ~/Desktop/shell-lesson-dataThen
cdinto theexercise-data/creaturesdirectory$ cd exercise-data/creaturesNow if you run
$ cd -you’ll see you’re back in
~/Desktop/shell-lesson-data. Runcd -again and you’re back in~/Desktop/shell-lesson-data/exercise-data/creatures
Absolute vs Relative Paths
Starting from
/Users/amanda/data, which of the following commands could Amanda use to navigate to her home directory, which is/Users/amanda?
cd .cd /cd /home/amandacd ../..cd ~cd homecd ~/data/..cdcd ..Solution
- No:
.stands for the current directory.- No:
/stands for the root directory.- No: Amanda’s home directory is
/Users/amanda.- No: this command goes up two levels, i.e. ends in
/Users.- Yes:
~stands for the user’s home directory, in this case/Users/amanda.- No: this command would navigate into a directory
homein the current directory if it exists.- Yes: unnecessarily complicated, but correct.
- Yes: shortcut to go back to the user’s home directory.
- Yes: goes up one level.
Relative Path Resolution
Using the filesystem diagram below, if
pwddisplays/Users/thing, what willls -F ../backupdisplay?
../backup: No such file or directory2012-12-01 2013-01-08 2013-01-272012-12-01/ 2013-01-08/ 2013-01-27/original/ pnas_final/ pnas_sub/
Solution
- No: there is a directory
backupin/Users.- No: this is the content of
Users/thing/backup, but with.., we asked for one level further up.- No: see previous explanation.
- Yes:
../backup/refers to/Users/backup/.
lsReading ComprehensionUsing the filesystem diagram below, if
pwddisplays/Users/backup, and-rtellslsto display things in reverse order, what command(s) will result in the following output:pnas_sub/ pnas_final/ original/
ls pwdls -r -Fls -r -F /Users/backupSolution
- No:
pwdis not the name of a directory.- Yes:
lswithout directory argument lists files and directories in the current directory.- Yes: uses the absolute path explicitly.
General Syntax of a Shell Command
We have now encountered commands, options, and arguments, but it is perhaps useful to formalise some terminology.
Consider the command below as a general example of a command, which we will dissect into its component parts:
$ ls -F /
ls is the command, with an option -F and an
argument /.
We’ve already encountered options which
either start with a single dash (-) or two dashes (--),
and they change the behavior of a command.
Arguments tell the command what to operate on (e.g. files and directories).
Sometimes options and arguments are referred to as parameters.
A command can be called with more than one option and more than one argument, but a
command doesn’t always require an argument or an option.
You might sometimes see options being referred to as switches or flags, especially for options that take no argument. In this lesson we will stick with using the term option.
Each part is separated by spaces: if you omit the space
between ls and -F the shell will look for a command called ls-F, which
doesn’t exist. Also, capitalization can be important.
For example, ls -s will display the size of files and directories alongside the names,
while ls -S will sort the files and directories by size, as shown below:
$ cd ~/Desktop/shell-lesson-data
$ ls -s exercise-data
total 28
4 animal-counts 4 creatures 12 numbers.txt 4 proteins 4 writing
$ ls -S exercise-data
animal-counts creatures proteins writing numbers.txt
Putting all that together, our command above gives us a listing
of files and directories in the root directory /.
An example of the output you might get from the above command is given below:
$ ls -F /
Applications/ System/
Library/ Users/
Network/ Volumes/
Nelle’s Pipeline: Organizing Files
Knowing this much about files and directories, Nelle is ready to organize the files that the protein assay machine will create.
She creates a directory called north-pacific-gyre
(to remind herself where the data came from),
which will contain the data files from the assay machine,
and her data processing scripts.
Each of her physical samples is labelled according to her lab’s convention
with a unique ten-character ID,
such as ‘NENE01729A’.
This ID is what she used in her collection log
to record the location, time, depth, and other characteristics of the sample,
so she decides to use it as part of each data file’s name.
Since the assay machine’s output is plain text,
she will call her files NENE01729A.txt, NENE01812A.txt, and so on.
All 1520 files will go into the same directory.
Now in her current directory shell-lesson-data,
Nelle can see what files she has using the command:
$ ls north-pacific-gyre/
This command is a lot to type, but she can let the shell do most of the work through what is called tab completion. If she types:
$ ls nor
and then presses Tab (the tab key on her keyboard), the shell automatically completes the directory name for her:
$ ls north-pacific-gyre/
Pressing Tab again does nothing, since there are multiple possibilities; pressing Tab twice brings up a list of all the files.
If Nelle adds G and presses Tab again, the shell will append ‘goo’ since all files that start with ‘g’ share the first three characters ‘goo’.
$ ls north-pacific-gyre/goo
To see all of those files, she can press Tab twice more.
ls north-pacific-gyre/goo
goodiff.sh goostats.sh
This is called tab completion, and we will see it in many other tools as we go on.
Key Points
The file system is responsible for managing information on the disk.
Information is stored in files, which are stored in directories (folders).
Directories can also store other directories, which then form a directory tree.
cd [path]changes the current working directory.
ls [path]prints a listing of a specific file or directory;lson its own lists the current working directory.
pwdprints the user’s current working directory.
/on its own is the root directory of the whole file system.Most commands take options that begin with a
-.A relative path specifies a location starting from the current location.
An absolute path specifies a location from the root of the file system.
Directory names in a path are separated with
/on Unix, but\on Windows.
..means ‘the directory above the current one’;.on its own means ‘the current directory’.
Working With Files and Directories
Overview
Teaching: 30 min
Exercises: 20 minQuestions
How can I create, copy, and delete files and directories?
How can I edit files?
Objectives
Create a directory hierarchy that matches a given diagram.
Create files in that hierarchy using an editor or by copying and renaming existing files.
Delete, copy and move specified files and/or directories.
Creating directories
We now know how to explore files and directories, but how do we create them in the first place?
In this episode we will learn about creating and moving files and directories,
using the exercise-data/writing directory as an example.
Step one: see where we are and what we already have
We should still be in the shell-lesson-data directory on the Desktop,
which we can checking using:
$ pwd
/Users/nelle/Desktop/shell-lesson-data
Next we’ll move to the exercise-data/writing directory and see what it contains:
$ cd exercise-data/writing/
$ ls -F
haiku.txt LittleWomen.txt
Create a directory
Let’s create a new directory called thesis using the command mkdir thesis
(which has no output):
$ mkdir thesis
As you might guess from its name,
mkdir means ‘make directory’.
Since thesis is a relative path
(i.e., does not have a leading slash, like /what/ever/thesis),
the new directory is created in the current working directory:
$ ls -F
haiku.txt LittleWomen.txt thesis/
Since we’ve just created the thesis directory, there’s nothing in it yet:
$ ls -F thesis
Note that mkdir is not limited to creating single directories one at a time.
The -p option allows mkdir to create a directory with nested subdirectories
in a single operation:
$ mkdir -p ../project/data ../project/results
The -R option to the ls command will list all nested subdirectories within a directory.
Let’s use ls -FR to recursively list the new directory hierarchy we just created in the
project directory:
$ ls -FR ../project
../project/:
data/ results/
../project/data:
../project/results:
Two ways of doing the same thing
Using the shell to create a directory is no different than using a file explorer. If you open the current directory using your operating system’s graphical file explorer, the
thesisdirectory will appear there too. While the shell and the file explorer are two different ways of interacting with the files, the files and directories themselves are the same.
Good names for files and directories
Complicated names of files and directories can make your life painful when working on the command line. Here we provide a few useful tips for the names of your files and directories.
Don’t use spaces.
Spaces can make a name more meaningful, but since spaces are used to separate arguments on the command line it is better to avoid them in names of files and directories. You can use
-or_instead (e.g.north-pacific-gyre/rather thannorth pacific gyre/). To test this out, try typingmkdir north pacific gyreand see what directory (or directories!) are made when you check withls -F.Don’t begin the name with
-(dash).Commands treat names starting with
-as options.Stick with letters, numbers,
.(period or ‘full stop’),-(dash) and_(underscore).Many other characters have special meanings on the command line. We will learn about some of these during this lesson. There are special characters that can cause your command to not work as expected and can even result in data loss.
If you need to refer to names of files or directories that have spaces or other special characters, you should surround the name in quotes (
"").
Create a text file
Let’s change our working directory to thesis using cd,
then run a text editor called Nano to create a file called draft.txt:
$ cd thesis
$ nano draft.txt
Which Editor?
When we say, ‘
nanois a text editor’ we really do mean ‘text’: it can only work with plain character data, not tables, images, or any other human-friendly media. We use it in examples because it is one of the least complex text editors. However, because of this trait, it may not be powerful enough or flexible enough for the work you need to do after this workshop. On Unix systems (such as Linux and macOS), many programmers use Emacs or Vim (both of which require more time to learn), or a graphical editor such as Gedit. On Windows, you may wish to use Notepad++. Windows also has a built-in editor callednotepadthat can be run from the command line in the same way asnanofor the purposes of this lesson.No matter what editor you use, you will need to know where it searches for and saves files. If you start it from the shell, it will (probably) use your current working directory as its default location. If you use your computer’s start menu, it may want to save files in your desktop or documents directory instead. You can change this by navigating to another directory the first time you ‘Save As…’
Let’s type in a few lines of text.
Once we’re happy with our text, we can press Ctrl+O
(press the Ctrl or Control key and, while
holding it down, press the O key) to write our data to disk
(we’ll be asked what file we want to save this to:
press Return to accept the suggested default of draft.txt).
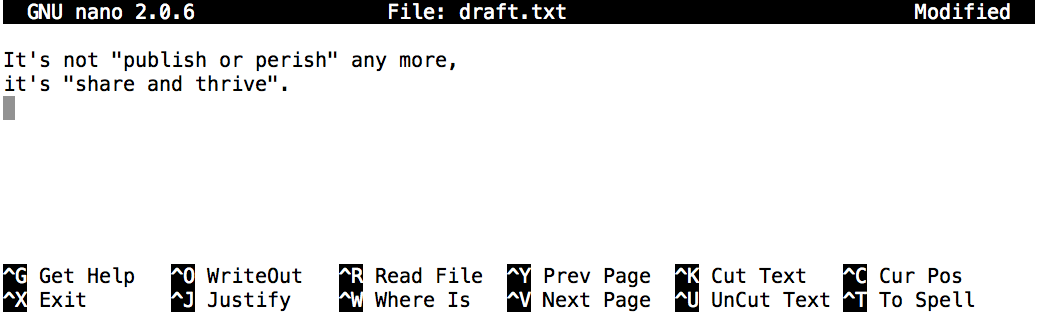
Once our file is saved, we can use Ctrl+X to quit the editor and return to the shell.
Control, Ctrl, or ^ Key
The Control key is also called the ‘Ctrl’ key. There are various ways in which using the Control key may be described. For example, you may see an instruction to press the Control key and, while holding it down, press the X key, described as any of:
Control-XControl+XCtrl-XCtrl+X^XC-xIn nano, along the bottom of the screen you’ll see
^G Get Help ^O WriteOut. This means that you can useControl-Gto get help andControl-Oto save your file.
nano doesn’t leave any output on the screen after it exits,
but ls now shows that we have created a file called draft.txt:
$ ls
draft.txt
Creating Files a Different Way
We have seen how to create text files using the
nanoeditor. Now, try the following command:$ touch my_file.txt
What did the
touchcommand do? When you look at your current directory using the GUI file explorer, does the file show up?Use
ls -lto inspect the files. How large ismy_file.txt?When might you want to create a file this way?
Solution
The
touchcommand generates a new file calledmy_file.txtin your current directory. You can observe this newly generated file by typinglsat the command line prompt.my_file.txtcan also be viewed in your GUI file explorer.When you inspect the file with
ls -l, note that the size ofmy_file.txtis 0 bytes. In other words, it contains no data. If you openmy_file.txtusing your text editor it is blank.Some programs do not generate output files themselves, but instead require that empty files have already been generated. When the program is run, it searches for an existing file to populate with its output. The touch command allows you to efficiently generate a blank text file to be used by such programs.
What’s In A Name?
You may have noticed that all of Nelle’s files are named ‘something dot something’, and in this part of the lesson, we always used the extension
.txt. This is just a convention: we can call a filemythesisor almost anything else we want. However, most people use two-part names most of the time to help them (and their programs) tell different kinds of files apart. The second part of such a name is called the filename extension and indicates what type of data the file holds:.txtsignals a plain text file,.cfgis a configuration file full of parameters for some program or other,.pngis a PNG image, and so on.This is just a convention, albeit an important one. Files contain bytes: it’s up to us and our programs to interpret those bytes according to the rules for plain text files, PDF documents, configuration files, images, and so on.
Naming a PNG image of a whale as
whale.mp3doesn’t somehow magically turn it into a recording of whale song, though it might cause the operating system to try to open it with a music player when someone double-clicks it.
Moving files and directories
Returning to the shell-lesson-data/writing directory,
$ cd ~/Desktop/shell-lesson-data/writing
In our thesis directory we have a file draft.txt
which isn’t a particularly informative name,
so let’s change the file’s name using mv,
which is short for ‘move’:
$ mv thesis/draft.txt thesis/quotes.txt
The first argument tells mv what we’re ‘moving’,
while the second is where it’s to go.
In this case,
we’re moving thesis/draft.txt to thesis/quotes.txt,
which has the same effect as renaming the file.
Sure enough,
ls shows us that thesis now contains one file called quotes.txt:
$ ls thesis
quotes.txt
One must be careful when specifying the target file name, since mv will
silently overwrite any existing file with the same name, which could
lead to data loss. An additional option, mv -i (or mv --interactive),
can be used to make mv ask you for confirmation before overwriting.
Note that mv also works on directories.
Let’s move quotes.txt into the current working directory.
We use mv once again,
but this time we’ll use just the name of a directory as the second argument
to tell mv that we want to keep the filename
but put the file somewhere new.
(This is why the command is called ‘move’.)
In this case,
the directory name we use is the special directory name . that we mentioned earlier.
$ mv thesis/quotes.txt .
The effect is to move the file from the directory it was in to the current working directory.
ls now shows us that thesis is empty:
$ ls thesis
$
Alternatively, we can confirm the file quotes.txt is no longer present in the thesis directory
by explicitly trying to list it:
$ ls thesis/quotes.txt
ls: cannot access 'thesis/quotes.txt': No such file or directory
ls with a filename or directory as an argument only lists the requested file or directory.
If the file given as the argument doesn’t exist, the shell returns an error as we saw above.
We can use this to see that quotes.txt is now present in our current directory:
$ ls quotes.txt
quotes.txt
Moving Files to a new folder
After running the following commands, Jamie realizes that she put the files
sucrose.datandmaltose.datinto the wrong folder. The files should have been placed in therawfolder.$ ls -F analyzed/ raw/ $ ls -F analyzed fructose.dat glucose.dat maltose.dat sucrose.dat $ cd analyzedFill in the blanks to move these files to the
raw/folder (i.e. the one she forgot to put them in)$ mv sucrose.dat maltose.dat ____/____Solution
$ mv sucrose.dat maltose.dat ../rawRecall that
..refers to the parent directory (i.e. one above the current directory) and that.refers to the current directory.
Copying files and directories
The cp command works very much like mv,
except it copies a file instead of moving it.
We can check that it did the right thing using ls
with two paths as arguments — like most Unix commands,
ls can be given multiple paths at once:
$ cp quotes.txt thesis/quotations.txt
$ ls quotes.txt thesis/quotations.txt
quotes.txt thesis/quotations.txt
We can also copy a directory and all its contents by using the
recursive option -r,
e.g. to back up a directory:
$ cp -r thesis thesis_backup
We can check the result by listing the contents of both the thesis and thesis_backup directory:
$ ls thesis thesis_backup
thesis:
quotations.txt
thesis_backup:
quotations.txt
Renaming Files
Suppose that you created a plain-text file in your current directory to contain a list of the statistical tests you will need to do to analyze your data, and named it:
statstics.txtAfter creating and saving this file you realize you misspelled the filename! You want to correct the mistake, which of the following commands could you use to do so?
cp statstics.txt statistics.txtmv statstics.txt statistics.txtmv statstics.txt .cp statstics.txt .Solution
- No. While this would create a file with the correct name, the incorrectly named file still exists in the directory and would need to be deleted.
- Yes, this would work to rename the file.
- No, the period(.) indicates where to move the file, but does not provide a new file name; identical file names cannot be created.
- No, the period(.) indicates where to copy the file, but does not provide a new file name; identical file names cannot be created.
Moving and Copying
What is the output of the closing
lscommand in the sequence shown below?$ pwd/Users/jamie/data$ lsproteins.dat$ mkdir recombined $ mv proteins.dat recombined/ $ cp recombined/proteins.dat ../proteins-saved.dat $ ls
proteins-saved.dat recombinedrecombinedproteins.dat recombinedproteins-saved.datSolution
We start in the
/Users/jamie/datadirectory, and create a new folder calledrecombined. The second line moves (mv) the fileproteins.datto the new folder (recombined). The third line makes a copy of the file we just moved. The tricky part here is where the file was copied to. Recall that..means ‘go up a level’, so the copied file is now in/Users/jamie. Notice that..is interpreted with respect to the current working directory, not with respect to the location of the file being copied. So, the only thing that will show using ls (in/Users/jamie/data) is the recombined folder.
- No, see explanation above.
proteins-saved.datis located at/Users/jamie- Yes
- No, see explanation above.
proteins.datis located at/Users/jamie/data/recombined- No, see explanation above.
proteins-saved.datis located at/Users/jamie
Removing files and directories
Returning to the shell-lesson-data/writing directory,
let’s tidy up this directory by removing the quotes.txt file we created.
The Unix command we’ll use for this is rm (short for ‘remove’):
$ rm quotes.txt
We can confirm the file has gone using ls:
$ ls quotes.txt
ls: cannot access 'quotes.txt': No such file or directory
Deleting Is Forever
The Unix shell doesn’t have a trash bin that we can recover deleted files from (though most graphical interfaces to Unix do). Instead, when we delete files, they are unlinked from the file system so that their storage space on disk can be recycled. Tools for finding and recovering deleted files do exist, but there’s no guarantee they’ll work in any particular situation, since the computer may recycle the file’s disk space right away.
Using
rmSafelyWhat happens when we execute
rm -i thesis_backup/quotations.txt? Why would we want this protection when usingrm?Solution
rm: remove regular file 'thesis_backup/quotations.txt'? yThe
-ioption will prompt before (every) removal (use Y to confirm deletion or N to keep the file). The Unix shell doesn’t have a trash bin, so all the files removed will disappear forever. By using the-ioption, we have the chance to check that we are deleting only the files that we want to remove.
If we try to remove the thesis directory using rm thesis,
we get an error message:
$ rm thesis
rm: cannot remove `thesis': Is a directory
This happens because rm by default only works on files, not directories.
rm can remove a directory and all its contents if we use the
recursive option -r, and it will do so without any confirmation prompts:
$ rm -r thesis
Given that there is no way to retrieve files deleted using the shell,
rm -r should be used with great caution
(you might consider adding the interactive option rm -r -i).
Operations with multiple files and directories
Oftentimes one needs to copy or move several files at once. This can be done by providing a list of individual filenames, or specifying a naming pattern using wildcards.
Copy with Multiple Filenames
For this exercise, you can test the commands in the
shell-lesson-data/exercise-datadirectory.In the example below, what does
cpdo when given several filenames and a directory name?$ mkdir backup $ cp creatures/minotaur.dat creatures/unicorn.dat backup/In the example below, what does
cpdo when given three or more file names?$ cd creatures $ ls -Fbasilisk.dat minotaur.dat unicorn.dat$ $ cp minotaur.dat unicorn.dat basilisk.datSolution
If given more than one file name followed by a directory name (i.e. the destination directory must be the last argument),
cpcopies the files to the named directory.If given three file names,
cpthrows an error such as the one below, because it is expecting a directory name as the last argument.cp: target 'basilisk.dat' is not a directory
Using wildcards for accessing multiple files at once
Wildcards
*is a wildcard, which matches zero or more characters. Let’s consider theshell-lesson-data/exercise-data/proteinsdirectory:*.pdbmatchesethane.pdb,propane.pdb, and every file that ends with ‘.pdb’. On the other hand,p*.pdbonly matchespentane.pdbandpropane.pdb, because the ‘p’ at the front only matches filenames that begin with the letter ‘p’.
?is also a wildcard, but it matches exactly one character. So?ethane.pdbwould matchmethane.pdbwhereas*ethane.pdbmatches bothethane.pdb, andmethane.pdb.Wildcards can be used in combination with each other e.g.
???ane.pdbmatches three characters followed byane.pdb, givingcubane.pdb ethane.pdb octane.pdb.When the shell sees a wildcard, it expands the wildcard to create a list of matching filenames before running the command that was asked for. As an exception, if a wildcard expression does not match any file, Bash will pass the expression as an argument to the command as it is. For example, typing
ls *.pdfin theproteinsdirectory (which contains only files with names ending with.pdb) results in an error message that there is no file calledwcandlssee the lists of file names matching these expressions, but not the wildcards themselves. It is the shell, not the other programs, that deals with expanding wildcards.
List filenames matching a pattern
When run in the
proteinsdirectory, whichlscommand(s) will produce this output?
ethane.pdb methane.pdb
ls *t*ane.pdbls *t?ne.*ls *t??ne.pdbls ethane.*Solution
The solution is
3.
1.shows all files whose names contain zero or more characters (*) followed by the lettert, then zero or more characters (*) followed byane.pdb. This givesethane.pdb methane.pdb octane.pdb pentane.pdb.
2.shows all files whose names start with zero or more characters (*) followed by the lettert, then a single character (?), thenne.followed by zero or more characters (*). This will give usoctane.pdbandpentane.pdbbut doesn’t match anything which ends inthane.pdb.
3.fixes the problems of option 2 by matching two characters (??) betweentandne. This is the solution.
4.only shows files starting withethane..
More on Wildcards
Sam has a directory containing calibration data, datasets, and descriptions of the datasets:
. ├── 2015-10-23-calibration.txt ├── 2015-10-23-dataset1.txt ├── 2015-10-23-dataset2.txt ├── 2015-10-23-dataset_overview.txt ├── 2015-10-26-calibration.txt ├── 2015-10-26-dataset1.txt ├── 2015-10-26-dataset2.txt ├── 2015-10-26-dataset_overview.txt ├── 2015-11-23-calibration.txt ├── 2015-11-23-dataset1.txt ├── 2015-11-23-dataset2.txt ├── 2015-11-23-dataset_overview.txt ├── backup │ ├── calibration │ └── datasets └── send_to_bob ├── all_datasets_created_on_a_23rd └── all_november_filesBefore heading off to another field trip, she wants to back up her data and send some datasets to her colleague Bob. Sam uses the following commands to get the job done:
$ cp *dataset* backup/datasets $ cp ____calibration____ backup/calibration $ cp 2015-____-____ send_to_bob/all_november_files/ $ cp ____ send_to_bob/all_datasets_created_on_a_23rd/Help Sam by filling in the blanks.
The resulting directory structure should look like this
. ├── 2015-10-23-calibration.txt ├── 2015-10-23-dataset1.txt ├── 2015-10-23-dataset2.txt ├── 2015-10-23-dataset_overview.txt ├── 2015-10-26-calibration.txt ├── 2015-10-26-dataset1.txt ├── 2015-10-26-dataset2.txt ├── 2015-10-26-dataset_overview.txt ├── 2015-11-23-calibration.txt ├── 2015-11-23-dataset1.txt ├── 2015-11-23-dataset2.txt ├── 2015-11-23-dataset_overview.txt ├── backup │ ├── calibration │ │ ├── 2015-10-23-calibration.txt │ │ ├── 2015-10-26-calibration.txt │ │ └── 2015-11-23-calibration.txt │ └── datasets │ ├── 2015-10-23-dataset1.txt │ ├── 2015-10-23-dataset2.txt │ ├── 2015-10-23-dataset_overview.txt │ ├── 2015-10-26-dataset1.txt │ ├── 2015-10-26-dataset2.txt │ ├── 2015-10-26-dataset_overview.txt │ ├── 2015-11-23-dataset1.txt │ ├── 2015-11-23-dataset2.txt │ └── 2015-11-23-dataset_overview.txt └── send_to_bob ├── all_datasets_created_on_a_23rd │ ├── 2015-10-23-dataset1.txt │ ├── 2015-10-23-dataset2.txt │ ├── 2015-10-23-dataset_overview.txt │ ├── 2015-11-23-dataset1.txt │ ├── 2015-11-23-dataset2.txt │ └── 2015-11-23-dataset_overview.txt └── all_november_files ├── 2015-11-23-calibration.txt ├── 2015-11-23-dataset1.txt ├── 2015-11-23-dataset2.txt └── 2015-11-23-dataset_overview.txtSolution
$ cp *calibration.txt backup/calibration $ cp 2015-11-* send_to_bob/all_november_files/ $ cp *-23-dataset* send_to_bob/all_datasets_created_on_a_23rd/
Organizing Directories and Files
Jamie is working on a project and she sees that her files aren’t very well organized:
$ ls -Fanalyzed/ fructose.dat raw/ sucrose.datThe
fructose.datandsucrose.datfiles contain output from her data analysis. What command(s) covered in this lesson does she need to run so that the commands below will produce the output shown?$ ls -Fanalyzed/ raw/$ ls analyzedfructose.dat sucrose.datSolution
mv *.dat analyzedJamie needs to move her files
fructose.datandsucrose.datto theanalyzeddirectory. The shell will expand *.dat to match all .dat files in the current directory. Themvcommand then moves the list of .dat files to the ‘analyzed’ directory.
Reproduce a folder structure
You’re starting a new experiment and would like to duplicate the directory structure from your previous experiment so you can add new data.
Assume that the previous experiment is in a folder called
2016-05-18, which contains adatafolder that in turn contains folders namedrawandprocessedthat contain data files. The goal is to copy the folder structure of the2016-05-18folder into a folder called2016-05-20so that your final directory structure looks like this:2016-05-20/ └── data ├── processed └── rawWhich of the following set of commands would achieve this objective? What would the other commands do?
$ mkdir 2016-05-20 $ mkdir 2016-05-20/data $ mkdir 2016-05-20/data/processed $ mkdir 2016-05-20/data/raw$ mkdir 2016-05-20 $ cd 2016-05-20 $ mkdir data $ cd data $ mkdir raw processed$ mkdir 2016-05-20/data/raw $ mkdir 2016-05-20/data/processed$ mkdir -p 2016-05-20/data/raw $ mkdir -p 2016-05-20/data/processed$ mkdir 2016-05-20 $ cd 2016-05-20 $ mkdir data $ mkdir raw processedSolution
The first two sets of commands achieve this objective. The first set uses relative paths to create the top-level directory before the subdirectories.
The third set of commands will give an error because the default behavior of
mkdirwon’t create a subdirectory of a non-existent directory: the intermediate level folders must be created first.The fourth set of commands achieve this objective. Remember, the
-poption, followed by a path of one or more directories, will causemkdirto create any intermediate subdirectories as required.The final set of commands generates the ‘raw’ and ‘processed’ directories at the same level as the ‘data’ directory.
Key Points
cp [old] [new]copies a file.
mkdir [path]creates a new directory.
mv [old] [new]moves (renames) a file or directory.
rm [path]removes (deletes) a file.
*matches zero or more characters in a filename, so*.txtmatches all files ending in.txt.
?matches any single character in a filename, so?.txtmatchesa.txtbut notany.txt.Use of the Control key may be described in many ways, including
Ctrl-X,Control-X, and^X.The shell does not have a trash bin: once something is deleted, it’s really gone.
Most files’ names are
something.extension. The extension isn’t required, and doesn’t guarantee anything, but is normally used to indicate the type of data in the file.Depending on the type of work you do, you may need a more powerful text editor than Nano.
Pipes and Filters
Overview
Teaching: 25 min
Exercises: 10 minQuestions
How can I combine existing commands to do new things?
Objectives
Redirect a command’s output to a file.
Construct command pipelines with two or more stages.
Explain what usually happens if a program or pipeline isn’t given any input to process.
Explain the advantage of linking commands with pipes and filters.
Now that we know a few basic commands,
we can finally look at the shell’s most powerful feature:
the ease with which it lets us combine existing programs in new ways.
We’ll start with the directory shell-lesson-data/exercise-data/proteins
that contains six files describing some simple organic molecules.
The .pdb extension indicates that these files are in Protein Data Bank format,
a simple text format that specifies the type and position of each atom in the molecule.
$ ls proteins
cubane.pdb methane.pdb pentane.pdb
ethane.pdb octane.pdb propane.pdb
Let’s go into that directory with cd and run an example command wc cubane.pdb:
$ cd proteins
$ wc cubane.pdb
20 156 1158 cubane.pdb
wc is the ‘word count’ command:
it counts the number of lines, words, and characters in files (from left to right, in that order).
If we run the command wc *.pdb, the * in *.pdb matches zero or more characters,
so the shell turns *.pdb into a list of all .pdb files in the current directory:
$ wc *.pdb
20 156 1158 cubane.pdb
12 84 622 ethane.pdb
9 57 422 methane.pdb
30 246 1828 octane.pdb
21 165 1226 pentane.pdb
15 111 825 propane.pdb
107 819 6081 total
Note that wc *.pdb also shows the total number of all lines in the last line of the output.
If we run wc -l instead of just wc,
the output shows only the number of lines per file:
$ wc -l *.pdb
20 cubane.pdb
12 ethane.pdb
9 methane.pdb
30 octane.pdb
21 pentane.pdb
15 propane.pdb
107 total
The -m and -w options can also be used with the wc command, to show
only the number of characters or the number of words in the files.
Why Isn’t It Doing Anything?
What happens if a command is supposed to process a file, but we don’t give it a filename? For example, what if we type:
$ wc -lbut don’t type
*.pdb(or anything else) after the command? Since it doesn’t have any filenames,wcassumes it is supposed to process input given at the command prompt, so it just sits there and waits for us to give it some data interactively. From the outside, though, all we see is it sitting there: the command doesn’t appear to do anything.If you make this kind of mistake, you can escape out of this state by holding down the control key (Ctrl) and typing the letter C once and letting go of the Ctrl key. Ctrl+C
Capturing output from commands
Which of these files contains the fewest lines? It’s an easy question to answer when there are only six files, but what if there were 6000? Our first step toward a solution is to run the command:
$ wc -l *.pdb > lengths.txt
The greater than symbol, >, tells the shell to redirect the command’s output
to a file instead of printing it to the screen. (This is why there is no screen output:
everything that wc would have printed has gone into the
file lengths.txt instead.) The shell will create
the file if it doesn’t exist. If the file exists, it will be
silently overwritten, which may lead to data loss and thus requires
some caution.
ls lengths.txt confirms that the file exists:
$ ls lengths.txt
lengths.txt
We can now send the content of lengths.txt to the screen using cat lengths.txt.
The cat command gets its name from ‘concatenate’ i.e. join together,
and it prints the contents of files one after another.
There’s only one file in this case,
so cat just shows us what it contains:
$ cat lengths.txt
20 cubane.pdb
12 ethane.pdb
9 methane.pdb
30 octane.pdb
21 pentane.pdb
15 propane.pdb
107 total
Output Page by Page
We’ll continue to use
catin this lesson, for convenience and consistency, but it has the disadvantage that it always dumps the whole file onto your screen. More useful in practice is the commandless, which you use withless lengths.txt. This displays a screenful of the file, and then stops. You can go forward one screenful by pressing the spacebar, or back one by pressingb. Pressqto quit.
Filtering output
Next we’ll use the sort command to sort the contents of the lengths.txt file.
But first we’ll use an exercise to learn a little about the sort command:
What Does
sort -nDo?The file
shell-lesson-data/exercise-data/numbers.txtcontains the following lines:10 2 19 22 6If we run
sorton this file, the output is:10 19 2 22 6If we run
sort -non the same file, we get this instead:2 6 10 19 22Explain why
-nhas this effect.Solution
The
-noption specifies a numerical rather than an alphanumerical sort.
We will also use the -n option to specify that the sort is
numerical instead of alphanumerical.
This does not change the file;
instead, it sends the sorted result to the screen:
$ sort -n lengths.txt
9 methane.pdb
12 ethane.pdb
15 propane.pdb
20 cubane.pdb
21 pentane.pdb
30 octane.pdb
107 total
We can put the sorted list of lines in another temporary file called sorted-lengths.txt
by putting > sorted-lengths.txt after the command,
just as we used > lengths.txt to put the output of wc into lengths.txt.
Once we’ve done that,
we can run another command called head to get the first few lines in sorted-lengths.txt:
$ sort -n lengths.txt > sorted-lengths.txt
$ head -n 1 sorted-lengths.txt
9 methane.pdb
Using -n 1 with head tells it that
we only want the first line of the file;
-n 20 would get the first 20,
and so on.
Since sorted-lengths.txt contains the lengths of our files ordered from least to greatest,
the output of head must be the file with the fewest lines.
Redirecting to the same file
It’s a very bad idea to try redirecting the output of a command that operates on a file to the same file. For example:
$ sort -n lengths.txt > lengths.txtDoing something like this may give you incorrect results and/or delete the contents of
lengths.txt.
What Does
>>Mean?We have seen the use of
>, but there is a similar operator>>which works slightly differently. We’ll learn about the differences between these two operators by printing some strings. We can use theechocommand to print strings e.g.$ echo The echo command prints textThe echo command prints textNow test the commands below to reveal the difference between the two operators:
$ echo hello > testfile01.txtand:
$ echo hello >> testfile02.txtHint: Try executing each command twice in a row and then examining the output files.
Solution
In the first example with
>, the string ‘hello’ is written totestfile01.txt, but the file gets overwritten each time we run the command.We see from the second example that the
>>operator also writes ‘hello’ to a file (in this casetestfile02.txt), but appends the string to the file if it already exists (i.e. when we run it for the second time).
Appending Data
We have already met the
headcommand, which prints lines from the start of a file.tailis similar, but prints lines from the end of a file instead.Consider the file
shell-lesson-data/exercise-data/animal-counts/animals.csv. After these commands, select the answer that corresponds to the fileanimals-subset.csv:$ head -n 3 animals.csv > animals-subset.csv $ tail -n 2 animals.csv >> animals-subset.csv
- The first three lines of
animals.csv- The last two lines of
animals.csv- The first three lines and the last two lines of
animals.csv- The second and third lines of
animals.csvSolution
Option 3 is correct. For option 1 to be correct we would only run the
headcommand. For option 2 to be correct we would only run thetailcommand. For option 4 to be correct we would have to pipe the output ofheadintotail -n 2by doinghead -n 3 animals.csv | tail -n 2 > animals-subset.csv
Passing output to another command
In our example of finding the file with the fewest lines,
we are using two intermediate files lengths.txt and sorted-lengths.txt to store output.
This is a confusing way to work because
even once you understand what wc, sort, and head do,
those intermediate files make it hard to follow what’s going on.
We can make it easier to understand by running sort and head together:
$ sort -n lengths.txt | head -n 1
9 methane.pdb
The vertical bar, |, between the two commands is called a pipe.
It tells the shell that we want to use
the output of the command on the left
as the input to the command on the right.
This has removed the need for the sorted-lengths.txt file.
Combining multiple commands
Nothing prevents us from chaining pipes consecutively.
We can for example send the output of wc directly to sort,
and then the resulting output to head.
This removes the need for any intermediate files.
We’ll start by using a pipe to send the output of wc to sort:
$ wc -l *.pdb | sort -n
9 methane.pdb
12 ethane.pdb
15 propane.pdb
20 cubane.pdb
21 pentane.pdb
30 octane.pdb
107 total
We can then send that output through another pipe, to head, so that the full pipeline becomes:
$ wc -l *.pdb | sort -n | head -n 1
9 methane.pdb
This is exactly like a mathematician nesting functions like log(3x)
and saying ‘the log of three times x’.
In our case,
the calculation is ‘head of sort of line count of *.pdb’.
The redirection and pipes used in the last few commands are illustrated below:
Piping Commands Together
In our current directory, we want to find the 3 files which have the least number of lines. Which command listed below would work?
wc -l * > sort -n > head -n 3wc -l * | sort -n | head -n 1-3wc -l * | head -n 3 | sort -nwc -l * | sort -n | head -n 3Solution
Option 4 is the solution. The pipe character
|is used to connect the output from one command to the input of another.>is used to redirect standard output to a file. Try it in theshell-lesson-data/exercise-data/proteinsdirectory!
Tools designed to work together
This idea of linking programs together is why Unix has been so successful.
Instead of creating enormous programs that try to do many different things,
Unix programmers focus on creating lots of simple tools that each do one job well,
and that work well with each other.
This programming model is called ‘pipes and filters’.
We’ve already seen pipes;
a filter is a program like wc or sort
that transforms a stream of input into a stream of output.
Almost all of the standard Unix tools can work this way:
unless told to do otherwise,
they read from standard input,
do something with what they’ve read,
and write to standard output.
The key is that any program that reads lines of text from standard input and writes lines of text to standard output can be combined with every other program that behaves this way as well. You can and should write your programs this way so that you and other people can put those programs into pipes to multiply their power.
Pipe Reading Comprehension
A file called
animals.csv(in theshell-lesson-data/exercise-data/animal-countsfolder) contains the following data:2012-11-05,deer,5 2012-11-05,rabbit,22 2012-11-05,raccoon,7 2012-11-06,rabbit,19 2012-11-06,deer,2 2012-11-06,fox,4 2012-11-07,rabbit,16 2012-11-07,bear,1 2012-11-05,deerWhat text passes through each of the pipes and the final redirect in the pipeline below?
$ cat animals.csv | head -n 5 | tail -n 3 | sort -r > final.txtHint: build the pipeline up one command at a time to test your understanding
Solution
The
headcommand extracts the first 5 lines fromanimals.csv. Then, the last 3 lines are extracted from the previous 5 by using thetailcommand. With thesort -rcommand those 3 lines are sorted in reverse order and finally, the output is redirected to a filefinal.txt. The content of this file can be checked by executingcat final.txt. The file should contain the following lines:2012-11-06,rabbit,19 2012-11-06,deer,2 2012-11-05,raccoon,7
Pipe Construction
For the file
animals.csvfrom the previous exercise, consider the following command:$ cut -d , -f 2 animals.csvThe
cutcommand is used to remove or ‘cut out’ certain sections of each line in the file, andcutexpects the lines to be separated into columns by a Tab character. A character used in this way is a called a delimiter. In the example above we use the-doption to specify the comma as our delimiter character. We have also used the-foption to specify that we want to extract the second field (column). This gives the following output:deer rabbit raccoon rabbit deer fox rabbit bearThe
uniqcommand filters out adjacent matching lines in a file. How could you extend this pipeline (usinguniqand another command) to find out what animals the file contains (without any duplicates in their names)?Solution
$ cut -d , -f 2 animals.csv | sort | uniq
Which Pipe?
The file
animals.csvcontains 8 lines of data formatted as follows:2012-11-05,deer,5 2012-11-05,rabbit,22 2012-11-05,raccoon,7 2012-11-06,rabbit,19 ...The
uniqcommand has a-coption which gives a count of the number of times a line occurs in its input. Assuming your current directory isshell-lesson-data/exercise-data/, what command would you use to produce a table that shows the total count of each type of animal in the file?
sort animals.csv | uniq -csort -t, -k2,2 animals.csv | uniq -ccut -d, -f 2 animals.csv | uniq -ccut -d, -f 2 animals.csv | sort | uniq -ccut -d, -f 2 animals.csv | sort | uniq -c | wc -lSolution
Option 4. is the correct answer. If you have difficulty understanding why, try running the commands, or sub-sections of the pipelines (make sure you are in the
shell-lesson-data/datadirectory).
Nelle’s Pipeline: Checking Files
Nelle has run her samples through the assay machines
and created 17 files in the north-pacific-gyre directory described earlier.
As a quick check, starting from the shell-lesson-data directory, Nelle types:
$ cd north-pacific-gyre
$ wc -l *.txt
The output is 18 lines that look like this:
300 NENE01729A.txt
300 NENE01729B.txt
300 NENE01736A.txt
300 NENE01751A.txt
300 NENE01751B.txt
300 NENE01812A.txt
... ...
Now she types this:
$ wc -l *.txt | sort -n | head -n 5
240 NENE02018B.txt
300 NENE01729A.txt
300 NENE01729B.txt
300 NENE01736A.txt
300 NENE01751A.txt
Whoops: one of the files is 60 lines shorter than the others. When she goes back and checks it, she sees that she did that assay at 8:00 on a Monday morning — someone was probably in using the machine on the weekend, and she forgot to reset it. Before re-running that sample, she checks to see if any files have too much data:
$ wc -l *.txt | sort -n | tail -n 5
300 NENE02040B.txt
300 NENE02040Z.txt
300 NENE02043A.txt
300 NENE02043B.txt
5040 total
Those numbers look good — but what’s that ‘Z’ doing there in the third-to-last line? All of her samples should be marked ‘A’ or ‘B’; by convention, her lab uses ‘Z’ to indicate samples with missing information. To find others like it, she does this:
$ ls *Z.txt
NENE01971Z.txt NENE02040Z.txt
Sure enough,
when she checks the log on her laptop,
there’s no depth recorded for either of those samples.
Since it’s too late to get the information any other way,
she must exclude those two files from her analysis.
She could delete them using rm,
but there are actually some analyses she might do later where depth doesn’t matter,
so instead, she’ll have to be careful later on to select files using the wildcard expressions
NENE*A.txt NENE*B.txt.
Removing Unneeded Files
Suppose you want to delete your processed data files, and only keep your raw files and processing script to save storage. The raw files end in
.datand the processed files end in.txt. Which of the following would remove all the processed data files, and only the processed data files?
rm ?.txtrm *.txtrm * .txtrm *.*Solution
- This would remove
.txtfiles with one-character names- This is correct answer
- The shell would expand
*to match everything in the current directory, so the command would try to remove all matched files and an additional file called.txt- The shell would expand
*.*to match all files with any extension, so this command would delete all files
Key Points
wccounts lines, words, and characters in its inputs.
catdisplays the contents of its inputs.
sortsorts its inputs.
headdisplays the first 10 lines of its input.
taildisplays the last 10 lines of its input.
command > [file]redirects a command’s output to a file (overwriting any existing content).
command >> [file]appends a command’s output to a file.
[first] | [second]is a pipeline: the output of the first command is used as the input to the second.The best way to use the shell is to use pipes to combine simple single-purpose programs (filters).
Loops
Overview
Teaching: 40 min
Exercises: 10 minQuestions
How can I perform the same actions on many different files?
Objectives
Write a loop that applies one or more commands separately to each file in a set of files.
Trace the values taken on by a loop variable during execution of the loop.
Explain the difference between a variable’s name and its value.
Explain why spaces and some punctuation characters shouldn’t be used in file names.
Demonstrate how to see what commands have recently been executed.
Re-run recently executed commands without retyping them.
Loops are a programming construct which allow us to repeat a command or set of commands for each item in a list. As such they are key to productivity improvements through automation. Similar to wildcards and tab completion, using loops also reduces the amount of typing required (and hence reduces the number of typing mistakes).
Suppose we have several hundred genome data files named basilisk.dat, minotaur.dat, and
unicorn.dat.
For this example, we’ll use the exercise-data/creatures directory which only has three
example files,
but the principles can be applied to many many more files at once.
The structure of these files is the same: the common name, classification, and updated date are presented on the first three lines, with DNA sequences on the following lines. Let’s look at the files:
$ head -n 5 basilisk.dat minotaur.dat unicorn.dat
We would like to print out the classification for each species, which is given on the second
line of each file.
For each file, we would need to execute the command head -n 2 and pipe this to tail -n 1.
We’ll use a loop to solve this problem, but first let’s look at the general form of a loop:
for thing in list_of_things
do
operation_using $thing # Indentation within the loop is not required, but aids legibility
done
and we can apply this to our example like this:
$ for filename in basilisk.dat minotaur.dat unicorn.dat
> do
> head -n 2 $filename | tail -n 1
> done
CLASSIFICATION: basiliscus vulgaris
CLASSIFICATION: bos hominus
CLASSIFICATION: equus monoceros
Follow the Prompt
The shell prompt changes from
$to>and back again as we were typing in our loop. The second prompt,>, is different to remind us that we haven’t finished typing a complete command yet. A semicolon,;, can be used to separate two commands written on a single line.
When the shell sees the keyword for,
it knows to repeat a command (or group of commands) once for each item in a list.
Each time the loop runs (called an iteration), an item in the list is assigned in sequence to
the variable, and the commands inside the loop are executed, before moving on to
the next item in the list.
Inside the loop,
we call for the variable’s value by putting $ in front of it.
The $ tells the shell interpreter to treat
the variable as a variable name and substitute its value in its place,
rather than treat it as text or an external command.
In this example, the list is three filenames: basilisk.dat, minotaur.dat, and unicorn.dat.
Each time the loop iterates, it will assign a file name to the variable filename
and run the head command.
The first time through the loop,
$filename is basilisk.dat.
The interpreter runs the command head on basilisk.dat
and pipes the first two lines to the tail command,
which then prints the second line of basilisk.dat.
For the second iteration, $filename becomes
minotaur.dat. This time, the shell runs head on minotaur.dat
and pipes the first two lines to the tail command,
which then prints the second line of minotaur.dat.
For the third iteration, $filename becomes
unicorn.dat, so the shell runs the head command on that file,
and tail on the output of that.
Since the list was only three items, the shell exits the for loop.
Same Symbols, Different Meanings
Here we see
>being used as a shell prompt, whereas>is also used to redirect output. Similarly,$is used as a shell prompt, but, as we saw earlier, it is also used to ask the shell to get the value of a variable.If the shell prints
>or$then it expects you to type something, and the symbol is a prompt.If you type
>or$yourself, it is an instruction from you that the shell should redirect output or get the value of a variable.
When using variables it is also
possible to put the names into curly braces to clearly delimit the variable
name: $filename is equivalent to ${filename}, but is different from
${file}name. You may find this notation in other people’s programs.
We have called the variable in this loop filename
in order to make its purpose clearer to human readers.
The shell itself doesn’t care what the variable is called;
if we wrote this loop as:
$ for x in basilisk.dat minotaur.dat unicorn.dat
> do
> head -n 2 $x | tail -n 1
> done
or:
$ for temperature in basilisk.dat minotaur.dat unicorn.dat
> do
> head -n 2 $temperature | tail -n 1
> done
it would work exactly the same way.
Don’t do this.
Programs are only useful if people can understand them,
so meaningless names (like x) or misleading names (like temperature)
increase the odds that the program won’t do what its readers think it does.
In the above examples, the variables (thing, filename, x and temperature)
could have been given any other name, as long as it is meaningful to both the person
writing the code and the person reading it.
Note also that loops can be used for other things than filenames, like a list of numbers or a subset of data.
Write your own loop
How would you write a loop that echoes all 10 numbers from 0 to 9?
Solution
$ for loop_variable in 0 1 2 3 4 5 6 7 8 9 > do > echo $loop_variable > done0 1 2 3 4 5 6 7 8 9
Variables in Loops
This exercise refers to the
shell-lesson-data/exercise-data/proteinsdirectory.ls *.pdbgives the following output:cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdbWhat is the output of the following code?
$ for datafile in *.pdb > do > ls *.pdb > doneNow, what is the output of the following code?
$ for datafile in *.pdb > do > ls $datafile > doneWhy do these two loops give different outputs?
Solution
The first code block gives the same output on each iteration through the loop. Bash expands the wildcard
*.pdbwithin the loop body (as well as before the loop starts) to match all files ending in.pdband then lists them usingls. The expanded loop would look like this:$ for datafile in cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb > do > ls cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb > donecubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdbThe second code block lists a different file on each loop iteration. The value of the
datafilevariable is evaluated using$datafile, and then listed usingls.cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb
Limiting Sets of Files
What would be the output of running the following loop in the
shell-lesson-data/exercise-data/proteinsdirectory?$ for filename in c* > do > ls $filename > done
- No files are listed.
- All files are listed.
- Only
cubane.pdb,octane.pdbandpentane.pdbare listed.- Only
cubane.pdbis listed.Solution
4 is the correct answer.
*matches zero or more characters, so any file name starting with the letter c, followed by zero or more other characters will be matched.How would the output differ from using this command instead?
$ for filename in *c* > do > ls $filename > done
- The same files would be listed.
- All the files are listed this time.
- No files are listed this time.
- The files
cubane.pdbandoctane.pdbwill be listed.- Only the file
octane.pdbwill be listed.Solution
4 is the correct answer.
*matches zero or more characters, so a file name with zero or more characters before a letter c and zero or more characters after the letter c will be matched.
Saving to a File in a Loop - Part One
In the
shell-lesson-data/exercise-data/proteinsdirectory, what is the effect of this loop?for alkanes in *.pdb do echo $alkanes cat $alkanes > alkanes.pdb done
- Prints
cubane.pdb,ethane.pdb,methane.pdb,octane.pdb,pentane.pdbandpropane.pdb, and the text frompropane.pdbwill be saved to a file calledalkanes.pdb.- Prints
cubane.pdb,ethane.pdb, andmethane.pdb, and the text from all three files would be concatenated and saved to a file calledalkanes.pdb.- Prints
cubane.pdb,ethane.pdb,methane.pdb,octane.pdb, andpentane.pdb, and the text frompropane.pdbwill be saved to a file calledalkanes.pdb.- None of the above.
Solution
- The text from each file in turn gets written to the
alkanes.pdbfile. However, the file gets overwritten on each loop iteration, so the final content ofalkanes.pdbis the text from thepropane.pdbfile.
Saving to a File in a Loop - Part Two
Also in the
shell-lesson-data/exercise-data/proteinsdirectory, what would be the output of the following loop?for datafile in *.pdb do cat $datafile >> all.pdb done
- All of the text from
cubane.pdb,ethane.pdb,methane.pdb,octane.pdb, andpentane.pdbwould be concatenated and saved to a file calledall.pdb.- The text from
ethane.pdbwill be saved to a file calledall.pdb.- All of the text from
cubane.pdb,ethane.pdb,methane.pdb,octane.pdb,pentane.pdbandpropane.pdbwould be concatenated and saved to a file calledall.pdb.- All of the text from
cubane.pdb,ethane.pdb,methane.pdb,octane.pdb,pentane.pdbandpropane.pdbwould be printed to the screen and saved to a file calledall.pdb.Solution
3 is the correct answer.
>>appends to a file, rather than overwriting it with the redirected output from a command. Given the output from thecatcommand has been redirected, nothing is printed to the screen.
Let’s continue with our example in the shell-lesson-data/exercise-data/creatures directory.
Here’s a slightly more complicated loop:
$ for filename in *.dat
> do
> echo $filename
> head -n 100 $filename | tail -n 20
> done
The shell starts by expanding *.dat to create the list of files it will process.
The loop body
then executes two commands for each of those files.
The first command, echo, prints its command-line arguments to standard output.
For example:
$ echo hello there
prints:
hello there
In this case,
since the shell expands $filename to be the name of a file,
echo $filename prints the name of the file.
Note that we can’t write this as:
$ for filename in *.dat
> do
> $filename
> head -n 100 $filename | tail -n 20
> done
because then the first time through the loop,
when $filename expanded to basilisk.dat, the shell would try to run basilisk.dat as
a program.
Finally,
the head and tail combination selects lines 81-100
from whatever file is being processed
(assuming the file has at least 100 lines).
Spaces in Names
Spaces are used to separate the elements of the list that we are going to loop over. If one of those elements contains a space character, we need to surround it with quotes, and do the same thing to our loop variable. Suppose our data files are named:
red dragon.dat purple unicorn.datTo loop over these files, we would need to add double quotes like so:
$ for filename in "red dragon.dat" "purple unicorn.dat" > do > head -n 100 "$filename" | tail -n 20 > doneIt is simpler to avoid using spaces (or other special characters) in filenames.
The files above don’t exist, so if we run the above code, the
headcommand will be unable to find them, however the error message returned will show the name of the files it is expecting:head: cannot open ‘red dragon.dat’ for reading: No such file or directory head: cannot open ‘purple unicorn.dat’ for reading: No such file or directoryTry removing the quotes around
$filenamein the loop above to see the effect of the quote marks on spaces. Note that we get a result from the loop command for unicorn.dat when we run this code in thecreaturesdirectory:head: cannot open ‘red’ for reading: No such file or directory head: cannot open ‘dragon.dat’ for reading: No such file or directory head: cannot open ‘purple’ for reading: No such file or directory CGGTACCGAA AAGGGTCGCG CAAGTGTTCC ...
We would like to modify each of the files in shell-lesson-data/exercise-data/creatures,
but also save a version
of the original files, naming the copies original-basilisk.dat and original-unicorn.dat.
We can’t use:
$ cp *.dat original-*.dat
because that would expand to:
$ cp basilisk.dat minotaur.dat unicorn.dat original-*.dat
This wouldn’t back up our files, instead we get an error:
cp: target `original-*.dat' is not a directory
This problem arises when cp receives more than two inputs. When this happens, it
expects the last input to be a directory where it can copy all the files it was passed.
Since there is no directory named original-*.dat in the creatures directory we get an
error.
Instead, we can use a loop:
$ for filename in *.dat
> do
> cp $filename original-$filename
> done
This loop runs the cp command once for each filename.
The first time,
when $filename expands to basilisk.dat,
the shell executes:
cp basilisk.dat original-basilisk.dat
The second time, the command is:
cp minotaur.dat original-minotaur.dat
The third and last time, the command is:
cp unicorn.dat original-unicorn.dat
Since the cp command does not normally produce any output, it’s hard to check
that the loop is doing the correct thing.
However, we learned earlier how to print strings using echo, and we can modify the loop
to use echo to print our commands without actually executing them.
As such we can check what commands would be run in the unmodified loop.
The following diagram
shows what happens when the modified loop is executed, and demonstrates how the
judicious use of echo is a good debugging technique.
Nelle’s Pipeline: Processing Files
Nelle is now ready to process her data files using goostats.sh —
a shell script written by her supervisor.
This calculates some statistics from a protein sample file, and takes two arguments:
- an input file (containing the raw data)
- an output file (to store the calculated statistics)
Since she’s still learning how to use the shell, she decides to build up the required commands in stages. Her first step is to make sure that she can select the right input files — remember, these are ones whose names end in ‘A’ or ‘B’, rather than ‘Z’. Starting from her home directory, Nelle types:
$ cd north-pacific-gyre
$ for datafile in NENE*A.txt NENE*B.txt
> do
> echo $datafile
> done
NENE01729A.txt
NENE01729B.txt
NENE01736A.txt
...
NENE02043A.txt
NENE02043B.txt
Her next step is to decide
what to call the files that the goostats.sh analysis program will create.
Prefixing each input file’s name with ‘stats’ seems simple,
so she modifies her loop to do that:
$ for datafile in NENE*A.txt NENE*B.txt
> do
> echo $datafile stats-$datafile
> done
NENE01729A.txt stats-NENE01729A.txt
NENE01729B.txt stats-NENE01729B.txt
NENE01736A.txt stats-NENE01736A.txt
...
NENE02043A.txt stats-NENE02043A.txt
NENE02043B.txt stats-NENE02043B.txt
She hasn’t actually run goostats.sh yet,
but now she’s sure she can select the right files and generate the right output filenames.
Typing in commands over and over again is becoming tedious, though, and Nelle is worried about making mistakes, so instead of re-entering her loop, she presses ↑. In response, the shell redisplays the whole loop on one line (using semi-colons to separate the pieces):
$ for datafile in NENE*A.txt NENE*B.txt; do echo $datafile stats-$datafile; done
Using the left arrow key,
Nelle backs up and changes the command echo to bash goostats.sh:
$ for datafile in NENE*A.txt NENE*B.txt; do bash goostats.sh $datafile stats-$datafile; done
When she presses Enter, the shell runs the modified command. However, nothing appears to happen — there is no output. After a moment, Nelle realizes that since her script doesn’t print anything to the screen any longer, she has no idea whether it is running, much less how quickly. She kills the running command by typing Ctrl+C, uses ↑ to repeat the command, and edits it to read:
$ for datafile in NENE*A.txt NENE*B.txt; do echo $datafile;
bash goostats.sh $datafile stats-$datafile; done
Beginning and End
We can move to the beginning of a line in the shell by typing Ctrl+A and to the end using Ctrl+E.
When she runs her program now, it produces one line of output every five seconds or so:
NENE01729A.txt
NENE01729B.txt
NENE01736A.txt
...
1518 times 5 seconds,
divided by 60,
tells her that her script will take about two hours to run.
As a final check,
she opens another terminal window,
goes into north-pacific-gyre,
and uses cat stats-NENE01729B.txt
to examine one of the output files.
It looks good,
so she decides to get some coffee and catch up on her reading.
Those Who Know History Can Choose to Repeat It
Another way to repeat previous work is to use the
historycommand to get a list of the last few hundred commands that have been executed, and then to use!123(where ‘123’ is replaced by the command number) to repeat one of those commands. For example, if Nelle types this:$ history | tail -n 5456 ls -l NENE0*.txt 457 rm stats-NENE01729B.txt.txt 458 bash goostats.sh NENE01729B.txt stats-NENE01729B.txt 459 ls -l NENE0*.txt 460 historythen she can re-run
goostats.shonNENE01729B.txtsimply by typing!458.
Other History Commands
There are a number of other shortcut commands for getting at the history.
- Ctrl+R enters a history search mode ‘reverse-i-search’ and finds the most recent command in your history that matches the text you enter next. Press Ctrl+R one or more additional times to search for earlier matches. You can then use the left and right arrow keys to choose that line and edit it then hit Return to run the command.
!!retrieves the immediately preceding command (you may or may not find this more convenient than using ↑)!$retrieves the last word of the last command. That’s useful more often than you might expect: afterbash goostats.sh NENE01729B.txt stats-NENE01729B.txt, you can typeless !$to look at the filestats-NENE01729B.txt, which is quicker than doing ↑ and editing the command-line.
Doing a Dry Run
A loop is a way to do many things at once — or to make many mistakes at once if it does the wrong thing. One way to check what a loop would do is to
echothe commands it would run instead of actually running them.Suppose we want to preview the commands the following loop will execute without actually running those commands:
$ for datafile in *.pdb > do > cat $datafile >> all.pdb > doneWhat is the difference between the two loops below, and which one would we want to run?
# Version 1 $ for datafile in *.pdb > do > echo cat $datafile >> all.pdb > done# Version 2 $ for datafile in *.pdb > do > echo "cat $datafile >> all.pdb" > doneSolution
The second version is the one we want to run. This prints to screen everything enclosed in the quote marks, expanding the loop variable name because we have prefixed it with a dollar sign. It also does not modify nor create the file
all.pdb, as the>>is treated literally as part of a string rather than as a redirection instruction.The first version appends the output from the command
echo cat $datafileto the file,all.pdb. This file will just contain the list;cat cubane.pdb,cat ethane.pdb,cat methane.pdbetc.Try both versions for yourself to see the output! Be sure to open the
all.pdbfile to view its contents.
Nested Loops
Suppose we want to set up a directory structure to organize some experiments measuring reaction rate constants with different compounds and different temperatures. What would be the result of the following code:
$ for species in cubane ethane methane > do > for temperature in 25 30 37 40 > do > mkdir $species-$temperature > done > doneSolution
We have a nested loop, i.e. contained within another loop, so for each species in the outer loop, the inner loop (the nested loop) iterates over the list of temperatures, and creates a new directory for each combination.
Try running the code for yourself to see which directories are created!
Key Points
A
forloop repeats commands once for every thing in a list.Every
forloop needs a variable to refer to the thing it is currently operating on.Use
$nameto expand a variable (i.e., get its value).${name}can also be used.Do not use spaces, quotes, or wildcard characters such as ‘*’ or ‘?’ in filenames, as it complicates variable expansion.
Give files consistent names that are easy to match with wildcard patterns to make it easy to select them for looping.
Use the up-arrow key to scroll up through previous commands to edit and repeat them.
Use Ctrl+R to search through the previously entered commands.
Use
historyto display recent commands, and![number]to repeat a command by number.
Shell Scripts
Overview
Teaching: 30 min
Exercises: 15 minQuestions
How can I save and re-use commands?
Objectives
Write a shell script that runs a command or series of commands for a fixed set of files.
Run a shell script from the command line.
Write a shell script that operates on a set of files defined by the user on the command line.
Create pipelines that include shell scripts you, and others, have written.
We are finally ready to see what makes the shell such a powerful programming environment. We are going to take the commands we repeat frequently and save them in files so that we can re-run all those operations again later by typing a single command. For historical reasons, a bunch of commands saved in a file is usually called a shell script, but make no mistake: these are actually small programs.
Not only will writing shell scripts make your work faster — you won’t have to retype the same commands over and over again — it will also make it more accurate (fewer chances for typos) and more reproducible. If you come back to your work later (or if someone else finds your work and wants to build on it) you will be able to reproduce the same results simply by running your script, rather than having to remember or retype a long list of commands.
Let’s start by going back to proteins/ and creating a new file, middle.sh which will
become our shell script:
$ cd proteins
$ nano middle.sh
The command nano middle.sh opens the file middle.sh within the text editor ‘nano’
(which runs within the shell).
If the file does not exist, it will be created.
We can use the text editor to directly edit the file – we’ll simply insert the following line:
head -n 15 octane.pdb | tail -n 5
This is a variation on the pipe we constructed earlier:
it selects lines 11-15 of the file octane.pdb.
Remember, we are not running it as a command just yet:
we are putting the commands in a file.
Then we save the file (Ctrl-O in nano),
and exit the text editor (Ctrl-X in nano).
Check that the directory proteins now contains a file called middle.sh.
Once we have saved the file,
we can ask the shell to execute the commands it contains.
Our shell is called bash, so we run the following command:
$ bash middle.sh
ATOM 9 H 1 -4.502 0.681 0.785 1.00 0.00
ATOM 10 H 1 -5.254 -0.243 -0.537 1.00 0.00
ATOM 11 H 1 -4.357 1.252 -0.895 1.00 0.00
ATOM 12 H 1 -3.009 -0.741 -1.467 1.00 0.00
ATOM 13 H 1 -3.172 -1.337 0.206 1.00 0.00
Sure enough, our script’s output is exactly what we would get if we ran that pipeline directly.
Text vs. Whatever
We usually call programs like Microsoft Word or LibreOffice Writer “text editors”, but we need to be a bit more careful when it comes to programming. By default, Microsoft Word uses
.docxfiles to store not only text, but also formatting information about fonts, headings, and so on. This extra information isn’t stored as characters and doesn’t mean anything to tools likehead: they expect input files to contain nothing but the letters, digits, and punctuation on a standard computer keyboard. When editing programs, therefore, you must either use a plain text editor, or be careful to save files as plain text.
What if we want to select lines from an arbitrary file?
We could edit middle.sh each time to change the filename,
but that would probably take longer than typing the command out again
in the shell and executing it with a new file name.
Instead, let’s edit middle.sh and make it more versatile:
$ nano middle.sh
Now, within “nano”, replace the text octane.pdb with the special variable called $1:
head -n 15 "$1" | tail -n 5
Inside a shell script,
$1 means ‘the first filename (or other argument) on the command line’.
We can now run our script like this:
$ bash middle.sh octane.pdb
ATOM 9 H 1 -4.502 0.681 0.785 1.00 0.00
ATOM 10 H 1 -5.254 -0.243 -0.537 1.00 0.00
ATOM 11 H 1 -4.357 1.252 -0.895 1.00 0.00
ATOM 12 H 1 -3.009 -0.741 -1.467 1.00 0.00
ATOM 13 H 1 -3.172 -1.337 0.206 1.00 0.00
or on a different file like this:
$ bash middle.sh pentane.pdb
ATOM 9 H 1 1.324 0.350 -1.332 1.00 0.00
ATOM 10 H 1 1.271 1.378 0.122 1.00 0.00
ATOM 11 H 1 -0.074 -0.384 1.288 1.00 0.00
ATOM 12 H 1 -0.048 -1.362 -0.205 1.00 0.00
ATOM 13 H 1 -1.183 0.500 -1.412 1.00 0.00
Double-Quotes Around Arguments
For the same reason that we put the loop variable inside double-quotes, in case the filename happens to contain any spaces, we surround
$1with double-quotes.
Currently, we need to edit middle.sh each time we want to adjust the range of
lines that is returned.
Let’s fix that by configuring our script to instead use three command-line arguments.
After the first command-line argument ($1), each additional argument that we
provide will be accessible via the special variables $1, $2, $3,
which refer to the first, second, third command-line arguments, respectively.
Knowing this, we can use additional arguments to define the range of lines to
be passed to head and tail respectively:
$ nano middle.sh
head -n "$2" "$1" | tail -n "$3"
We can now run:
$ bash middle.sh pentane.pdb 15 5
ATOM 9 H 1 1.324 0.350 -1.332 1.00 0.00
ATOM 10 H 1 1.271 1.378 0.122 1.00 0.00
ATOM 11 H 1 -0.074 -0.384 1.288 1.00 0.00
ATOM 12 H 1 -0.048 -1.362 -0.205 1.00 0.00
ATOM 13 H 1 -1.183 0.500 -1.412 1.00 0.00
By changing the arguments to our command we can change our script’s behaviour:
$ bash middle.sh pentane.pdb 20 5
ATOM 14 H 1 -1.259 1.420 0.112 1.00 0.00
ATOM 15 H 1 -2.608 -0.407 1.130 1.00 0.00
ATOM 16 H 1 -2.540 -1.303 -0.404 1.00 0.00
ATOM 17 H 1 -3.393 0.254 -0.321 1.00 0.00
TER 18 1
This works,
but it may take the next person who reads middle.sh a moment to figure out what it does.
We can improve our script by adding some comments at the top:
$ nano middle.sh
# Select lines from the middle of a file.
# Usage: bash middle.sh filename end_line num_lines
head -n "$2" "$1" | tail -n "$3"
A comment starts with a # character and runs to the end of the line.
The computer ignores comments,
but they’re invaluable for helping people (including your future self) understand and use scripts.
The only caveat is that each time you modify the script,
you should check that the comment is still accurate:
an explanation that sends the reader in the wrong direction is worse than none at all.
What if we want to process many files in a single pipeline?
For example, if we want to sort our .pdb files by length, we would type:
$ wc -l *.pdb | sort -n
because wc -l lists the number of lines in the files
(recall that wc stands for ‘word count’, adding the -l option means ‘count lines’ instead)
and sort -n sorts things numerically.
We could put this in a file,
but then it would only ever sort a list of .pdb files in the current directory.
If we want to be able to get a sorted list of other kinds of files,
we need a way to get all those names into the script.
We can’t use $1, $2, and so on
because we don’t know how many files there are.
Instead, we use the special variable $@,
which means,
‘All of the command-line arguments to the shell script’.
We also should put $@ inside double-quotes
to handle the case of arguments containing spaces
("$@" is special syntax and is equivalent to "$1" "$2" …).
Here’s an example:
$ nano sorted.sh
# Sort files by their length.
# Usage: bash sorted.sh one_or_more_filenames
wc -l "$@" | sort -n
$ bash sorted.sh *.pdb ../creatures/*.dat
9 methane.pdb
12 ethane.pdb
15 propane.pdb
20 cubane.pdb
21 pentane.pdb
30 octane.pdb
163 ../creatures/basilisk.dat
163 ../creatures/minotaur.dat
163 ../creatures/unicorn.dat
596 total
List Unique Species
Leah has several hundred data files, each of which is formatted like this:
2013-11-05,deer,5 2013-11-05,rabbit,22 2013-11-05,raccoon,7 2013-11-06,rabbit,19 2013-11-06,deer,2 2013-11-06,fox,1 2013-11-07,rabbit,18 2013-11-07,bear,1An example of this type of file is given in
shell-lesson-data/exercise-data/animal-counts/animals.csv.We can use the command
cut -d , -f 2 animals.txt | sort | uniqto produce the unique species inanimals.txt. In order to avoid having to type out this series of commands every time, a scientist may choose to write a shell script instead.Write a shell script called
species.shthat takes any number of filenames as command-line arguments, and uses a variation of the above command to print a list of the unique species appearing in each of those files separately.Solution
# Script to find unique species in csv files where species is the second data field # This script accepts any number of file names as command line arguments # Loop over all files for file in $@ do echo "Unique species in $file:" # Extract species names cut -d , -f 2 $file | sort | uniq done
Suppose we have just run a series of commands that did something useful — for example, that created a graph we’d like to use in a paper. We’d like to be able to re-create the graph later if we need to, so we want to save the commands in a file. Instead of typing them in again (and potentially getting them wrong) we can do this:
$ history | tail -n 5 > redo-figure-3.sh
The file redo-figure-3.sh now contains:
297 bash goostats.sh NENE01729B.txt stats-NENE01729B.txt
298 bash goodiff.sh stats-NENE01729B.txt /data/validated/01729.txt > 01729-differences.txt
299 cut -d ',' -f 2-3 01729-differences.txt > 01729-time-series.txt
300 ygraph --format scatter --color bw --borders none 01729-time-series.txt figure-3.png
301 history | tail -n 5 > redo-figure-3.sh
After a moment’s work in an editor to remove the serial numbers on the commands,
and to remove the final line where we called the history command,
we have a completely accurate record of how we created that figure.
Why Record Commands in the History Before Running Them?
If you run the command:
$ history | tail -n 5 > recent.shthe last command in the file is the
historycommand itself, i.e., the shell has addedhistoryto the command log before actually running it. In fact, the shell always adds commands to the log before running them. Why do you think it does this?Solution
If a command causes something to crash or hang, it might be useful to know what that command was, in order to investigate the problem. Were the command only be recorded after running it, we would not have a record of the last command run in the event of a crash.
In practice, most people develop shell scripts by running commands at the shell prompt a few times
to make sure they’re doing the right thing,
then saving them in a file for re-use.
This style of work allows people to recycle
what they discover about their data and their workflow with one call to history
and a bit of editing to clean up the output
and save it as a shell script.
Nelle’s Pipeline: Creating a Script
Nelle’s supervisor insisted that all her analytics must be reproducible. The easiest way to capture all the steps is in a script.
First we return to Nelle’s project directory:
$ cd ../../north-pacific-gyre/
She creates a file using nano …
$ nano do-stats.sh
…which contains the following:
# Calculate stats for data files.
for datafile in "$@"
do
echo $datafile
bash goostats.sh $datafile stats-$datafile
done
She saves this in a file called do-stats.sh
so that she can now re-do the first stage of her analysis by typing:
$ bash do-stats.sh NENE*A.txt NENE*B.txt
She can also do this:
$ bash do-stats.sh NENE*A.txt NENE*B.txt | wc -l
so that the output is just the number of files processed rather than the names of the files that were processed.
One thing to note about Nelle’s script is that it lets the person running it decide what files to process. She could have written it as:
# Calculate stats for Site A and Site B data files.
for datafile in NENE*A.txt NENE*B.txt
do
echo $datafile
bash goostats.sh $datafile stats-$datafile
done
The advantage is that this always selects the right files:
she doesn’t have to remember to exclude the ‘Z’ files.
The disadvantage is that it always selects just those files — she can’t run it on all files
(including the ‘Z’ files),
or on the ‘G’ or ‘H’ files her colleagues in Antarctica are producing,
without editing the script.
If she wanted to be more adventurous,
she could modify her script to check for command-line arguments,
and use NENE*A.txt NENE*B.txt if none were provided.
Of course, this introduces another tradeoff between flexibility and complexity.
Variables in Shell Scripts
In the
proteinsdirectory, imagine you have a shell script calledscript.shcontaining the following commands:head -n $2 $1 tail -n $3 $1While you are in the
proteinsdirectory, you type the following command:$ bash script.sh '*.pdb' 1 1Which of the following outputs would you expect to see?
- All of the lines between the first and the last lines of each file ending in
.pdbin theproteinsdirectory- The first and the last line of each file ending in
.pdbin theproteinsdirectory- The first and the last line of each file in the
proteinsdirectory- An error because of the quotes around
*.pdbSolution
The correct answer is 2.
The special variables $1, $2 and $3 represent the command line arguments given to the script, such that the commands run are:
$ head -n 1 cubane.pdb ethane.pdb octane.pdb pentane.pdb propane.pdb $ tail -n 1 cubane.pdb ethane.pdb octane.pdb pentane.pdb propane.pdbThe shell does not expand
'*.pdb'because it is enclosed by quote marks. As such, the first argument to the script is'*.pdb'which gets expanded within the script byheadandtail.
Find the Longest File With a Given Extension
Write a shell script called
longest.shthat takes the name of a directory and a filename extension as its arguments, and prints out the name of the file with the most lines in that directory with that extension. For example:$ bash longest.sh shell-lesson-data/data/pdb pdbwould print the name of the
.pdbfile inshell-lesson-data/data/pdbthat has the most lines.Feel free to test your script on another directory e.g.
$ bash longest.sh shell-lesson-data/writing/data txtSolution
# Shell script which takes two arguments: # 1. a directory name # 2. a file extension # and prints the name of the file in that directory # with the most lines which matches the file extension. wc -l $1/*.$2 | sort -n | tail -n 2 | head -n 1The first part of the pipeline,
wc -l $1/*.$2 | sort -n, counts the lines in each file and sorts them numerically (largest last). When there’s more than one file,wcalso outputs a final summary line, giving the total number of lines across all files. We usetail -n 2 | head -n 1to throw away this last line.With
wc -l $1/*.$2 | sort -n | tail -n 1we’ll see the final summary line: we can build our pipeline up in pieces to be sure we understand the output.
Script Reading Comprehension
For this question, consider the
shell-lesson-data/exercise-data/proteinsdirectory once again. This contains a number of.pdbfiles in addition to any other files you may have created. Explain what each of the following three scripts would do when run asbash script1.sh *.pdb,bash script2.sh *.pdb, andbash script3.sh *.pdbrespectively.# Script 1 echo *.*# Script 2 for filename in $1 $2 $3 do cat $filename done# Script 3 echo $@.pdbSolutions
In each case, the shell expands the wildcard in
*.pdbbefore passing the resulting list of file names as arguments to the script.Script 1 would print out a list of all files containing a dot in their name. The arguments passed to the script are not actually used anywhere in the script.
Script 2 would print the contents of the first 3 files with a
.pdbfile extension.$1,$2, and$3refer to the first, second, and third argument respectively.Script 3 would print all the arguments to the script (i.e. all the
.pdbfiles), followed by.pdb.$@refers to all the arguments given to a shell script.cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb.pdb
Debugging Scripts
Suppose you have saved the following script in a file called
do-errors.shin Nelle’snorth-pacific-gyre/scriptsdirectory:# Calculate stats for data files. for datafile in "$@" do echo $datfile bash goostats.sh $datafile stats-$datafile doneWhen you run it from the
north-pacific-gyredirectory:$ bash do-errors.sh NENE*A.txt NENE*B.txtthe output is blank. To figure out why, re-run the script using the
-xoption:$ bash -x do-errors.sh NENE*A.txt NENE*B.txtWhat is the output showing you? Which line is responsible for the error?
Solution
The
-xoption causesbashto run in debug mode. This prints out each command as it is run, which will help you to locate errors. In this example, we can see thatechoisn’t printing anything. We have made a typo in the loop variable name, and the variabledatfiledoesn’t exist, hence returning an empty string.
Key Points
Save commands in files (usually called shell scripts) for re-use.
bash [filename]runs the commands saved in a file.
$@refers to all of a shell script’s command-line arguments.
$1,$2, etc., refer to the first command-line argument, the second command-line argument, etc.Place variables in quotes if the values might have spaces in them.
Letting users decide what files to process is more flexible and more consistent with built-in Unix commands.
Finding Things
Overview
Teaching: 25 min
Exercises: 20 minQuestions
How can I find files?
How can I find things in files?
Objectives
Use
grepto select lines from text files that match simple patterns.Use
findto find files and directories whose names match simple patterns.Use the output of one command as the command-line argument(s) to another command.
Explain what is meant by ‘text’ and ‘binary’ files, and why many common tools don’t handle the latter well.
In the same way that many of us now use ‘Google’ as a verb meaning ‘to find’, Unix programmers often use the word ‘grep’. ‘grep’ is a contraction of ‘global/regular expression/print’, a common sequence of operations in early Unix text editors. It is also the name of a very useful command-line program.
grep finds and prints lines in files that match a pattern.
For our examples,
we will use a file that contains three haiku taken from a
1998 competition in Salon magazine. For this set of examples,
we’re going to be working in the writing subdirectory:
$ cd
$ cd Desktop/shell-lesson-data/exercise-data/writing
$ cat haiku.txt
The Tao that is seen
Is not the true Tao, until
You bring fresh toner.
With searching comes loss
and the presence of absence:
"My Thesis" not found.
Yesterday it worked
Today it is not working
Software is like that.
Forever, or Five Years
We haven’t linked to the original haiku because they don’t appear to be on Salon’s site any longer. As Jeff Rothenberg said, ‘Digital information lasts forever — or five years, whichever comes first.’ Luckily, popular content often has backups.
Let’s find lines that contain the word ‘not’:
$ grep not haiku.txt
Is not the true Tao, until
"My Thesis" not found
Today it is not working
Here, not is the pattern we’re searching for.
The grep command searches through the file, looking for matches to the pattern specified.
To use it type grep, then the pattern we’re searching for and finally
the name of the file (or files) we’re searching in.
The output is the three lines in the file that contain the letters ‘not’.
By default, grep searches for a pattern in a case-sensitive way. In addition, the search pattern we have selected does not have to form a complete word, as we will see in the next example.
Let’s search for the pattern: ‘The’.
$ grep The haiku.txt
The Tao that is seen
"My Thesis" not found.
This time, two lines that include the letters ‘The’ are outputted, one of which contained our search pattern within a larger word, ‘Thesis’.
To restrict matches to lines containing the word ‘The’ on its own,
we can give grep with the -w option.
This will limit matches to word boundaries.
Later in this lesson, we will also see how we can change the search behavior of grep with respect to its case sensitivity.
$ grep -w The haiku.txt
The Tao that is seen
Note that a ‘word boundary’ includes the start and end of a line, so not
just letters surrounded by spaces.
Sometimes we don’t
want to search for a single word, but a phrase. This is also easy to do with
grep by putting the phrase in quotes.
$ grep -w "is not" haiku.txt
Today it is not working
We’ve now seen that you don’t have to have quotes around single words, but it is useful to use quotes when searching for multiple words. It also helps to make it easier to distinguish between the search term or phrase and the file being searched. We will use quotes in the remaining examples.
Another useful option is -n, which numbers the lines that match:
$ grep -n "it" haiku.txt
5:With searching comes loss
9:Yesterday it worked
10:Today it is not working
Here, we can see that lines 5, 9, and 10 contain the letters ‘it’.
We can combine options (i.e. flags) as we do with other Unix commands.
For example, let’s find the lines that contain the word ‘the’.
We can combine the option -w to find the lines that contain the word ‘the’
and -n to number the lines that match:
$ grep -n -w "the" haiku.txt
2:Is not the true Tao, until
6:and the presence of absence:
Now we want to use the option -i to make our search case-insensitive:
$ grep -n -w -i "the" haiku.txt
1:The Tao that is seen
2:Is not the true Tao, until
6:and the presence of absence:
Now, we want to use the option -v to invert our search, i.e., we want to output
the lines that do not contain the word ‘the’.
$ grep -n -w -v "the" haiku.txt
1:The Tao that is seen
3:You bring fresh toner.
4:
5:With searching comes loss
7:"My Thesis" not found.
8:
9:Yesterday it worked
10:Today it is not working
11:Software is like that.
If we use the -r (recursive) option,
grep can search for a pattern recursively through a set of files in subdirectories.
Let’s search recursively for Yesterday in the shell-lesson-data/exercise-data/writing directory:
$ grep -r Yesterday .
./LittleWomen.txt:"Yesterday, when Aunt was asleep and I was trying to be as still as a
./LittleWomen.txt:Yesterday at dinner, when an Austrian officer stared at us and then
./LittleWomen.txt:Yesterday was a quiet day spent in teaching, sewing, and writing in my
./haiku.txt:Yesterday it worked
grep has lots of other options. To find out what they are, we can type:
$ grep --help
Usage: grep [OPTION]... PATTERN [FILE]...
Search for PATTERN in each FILE or standard input.
PATTERN is, by default, a basic regular expression (BRE).
Example: grep -i 'hello world' menu.h main.c
Regexp selection and interpretation:
-E, --extended-regexp PATTERN is an extended regular expression (ERE)
-F, --fixed-strings PATTERN is a set of newline-separated fixed strings
-G, --basic-regexp PATTERN is a basic regular expression (BRE)
-P, --perl-regexp PATTERN is a Perl regular expression
-e, --regexp=PATTERN use PATTERN for matching
-f, --file=FILE obtain PATTERN from FILE
-i, --ignore-case ignore case distinctions
-w, --word-regexp force PATTERN to match only whole words
-x, --line-regexp force PATTERN to match only whole lines
-z, --null-data a data line ends in 0 byte, not newline
Miscellaneous:
... ... ...
Using
grepWhich command would result in the following output:
and the presence of absence:
grep "of" haiku.txtgrep -E "of" haiku.txtgrep -w "of" haiku.txtgrep -i "of" haiku.txtSolution
The correct answer is 3, because the
-woption looks only for whole-word matches. The other options will also match ‘of’ when part of another word.
Wildcards
grep’s real power doesn’t come from its options, though; it comes from the fact that patterns can include wildcards. (The technical name for these is regular expressions, which is what the ‘re’ in ‘grep’ stands for.) Regular expressions are both complex and powerful; if you want to do complex searches, please look at the lesson on our website. As a taster, we can find lines that have an ‘o’ in the second position like this:$ grep -E "^.o" haiku.txtYou bring fresh toner. Today it is not working Software is like that.We use the
-Eoption and put the pattern in quotes to prevent the shell from trying to interpret it. (If the pattern contained a*, for example, the shell would try to expand it before runninggrep.) The^in the pattern anchors the match to the start of the line. The.matches a single character (just like?in the shell), while theomatches an actual ‘o’.
Tracking a Species
Leah has several hundred data files saved in one directory, each of which is formatted like this:
2013-11-05,deer,5 2013-11-05,rabbit,22 2013-11-05,raccoon,7 2013-11-06,rabbit,19 2013-11-06,deer,2She wants to write a shell script that takes a species as the first command-line argument and a directory as the second argument. The script should return one file called
species.txtcontaining a list of dates and the number of that species seen on each date. For example using the data shown above,rabbit.txtwould contain:2013-11-05,22 2013-11-06,19Put these commands and pipes in the right order to achieve this:
cut -d : -f 2 > | grep -w $1 -r $2 | $1.txt cut -d , -f 1,3Hint: use
man grepto look for how to grep text recursively in a directory andman cutto select more than one field in a line.An example of such a file is provided in
shell-lesson-data/exercise-data/animal-counts/animals.csvSolution
grep -w $1 -r $2 | cut -d : -f 2 | cut -d , -f 1,3 > $1.txtActually, you can swap the order of the two cut commands and it still works. At the command line, try changing the order of the cut commands, and have a look at the output from each step to see why this is the case.
You would call the script above like this:
$ bash count-species.sh bear .
Little Women
You and your friend, having just finished reading Little Women by Louisa May Alcott, are in an argument. Of the four sisters in the book, Jo, Meg, Beth, and Amy, your friend thinks that Jo was the most mentioned. You, however, are certain it was Amy. Luckily, you have a file
LittleWomen.txtcontaining the full text of the novel (shell-lesson-data/exercise-data/writing/LittleWomen.txt). Using aforloop, how would you tabulate the number of times each of the four sisters is mentioned?Hint: one solution might employ the commands
grepandwcand a|, while another might utilizegrepoptions. There is often more than one way to solve a programming task, so a particular solution is usually chosen based on a combination of yielding the correct result, elegance, readability, and speed.Solutions
for sis in Jo Meg Beth Amy do echo $sis: grep -ow $sis LittleWomen.txt | wc -l doneAlternative, slightly inferior solution:
for sis in Jo Meg Beth Amy do echo $sis: grep -ocw $sis LittleWomen.txt doneThis solution is inferior because
grep -conly reports the number of lines matched. The total number of matches reported by this method will be lower if there is more than one match per line.Perceptive observers may have noticed that character names sometimes appear in all-uppercase in chapter titles (e.g. ‘MEG GOES TO VANITY FAIR’). If you wanted to count these as well, you could add the
-ioption for case-insensitivity (though in this case, it doesn’t affect the answer to which sister is mentioned most frequently).
While grep finds lines in files,
the find command finds files themselves.
Again,
it has a lot of options;
to show how the simplest ones work, we’ll use the shell-lesson-data/exercise-data
directory tree shown below.
.
├── animal-counts/
│ └── animals.csv
├── creatures/
│ ├── basilisk.dat
│ ├── minotaur.dat
│ └── unicorn.dat
├── numbers.txt
├── proteins/
│ ├── cubane.pdb
│ ├── ethane.pdb
│ ├── methane.pdb
│ ├── octane.pdb
│ ├── pentane.pdb
│ └── propane.pdb
└── writing/
├── haiku.txt
└── LittleWomen.txt
The exercise-data directory contains one file, numbers.txt and four directories:
animal-counts, creatures, proteins and writing containing various files.
For our first command,
let’s run find . (remember to run this command from the shell-lesson-data/exercise-data folder).
$ find .
.
./writing
./writing/LittleWomen.txt
./writing/haiku.txt
./creatures
./creatures/basilisk.dat
./creatures/unicorn.dat
./creatures/minotaur.dat
./animal-counts
./animal-counts/animals.csv
./numbers.txt
./proteins
./proteins/ethane.pdb
./proteins/propane.pdb
./proteins/octane.pdb
./proteins/pentane.pdb
./proteins/methane.pdb
./proteins/cubane.pdb
As always, the . on its own means the current working directory,
which is where we want our search to start.
find’s output is the names of every file and directory
under the current working directory.
This can seem useless at first but find has many options
to filter the output and in this lesson we will discover some
of them.
The first option in our list is
-type d that means ‘things that are directories’.
Sure enough, find’s output is the names of the five directories (including .):
$ find . -type d
.
./writing
./creatures
./animal-counts
./proteins
Notice that the objects find finds are not listed in any particular order.
If we change -type d to -type f,
we get a listing of all the files instead:
$ find . -type f
./writing/LittleWomen.txt
./writing/haiku.txt
./creatures/basilisk.dat
./creatures/unicorn.dat
./creatures/minotaur.dat
./animal-counts/animals.csv
./numbers.txt
./proteins/ethane.pdb
./proteins/propane.pdb
./proteins/octane.pdb
./proteins/pentane.pdb
./proteins/methane.pdb
./proteins/cubane.pdb
Now let’s try matching by name:
$ find . -name *.txt
./numbers.txt
We expected it to find all the text files,
but it only prints out ./numbers.txt.
The problem is that the shell expands wildcard characters like * before commands run.
Since *.txt in the current directory expands to ./numbers.txt,
the command we actually ran was:
$ find . -name numbers.txt
find did what we asked; we just asked for the wrong thing.
To get what we want,
let’s do what we did with grep:
put *.txt in quotes to prevent the shell from expanding the * wildcard.
This way,
find actually gets the pattern *.txt, not the expanded filename numbers.txt:
$ find . -name "*.txt"
./writing/LittleWomen.txt
./writing/haiku.txt
./numbers.txt
Listing vs. Finding
lsandfindcan be made to do similar things given the right options, but under normal circumstances,lslists everything it can, whilefindsearches for things with certain properties and shows them.
As we said earlier,
the command line’s power lies in combining tools.
We’ve seen how to do that with pipes;
let’s look at another technique.
As we just saw,
find . -name "*.txt" gives us a list of all text files in or below the current directory.
How can we combine that with wc -l to count the lines in all those files?
The simplest way is to put the find command inside $():
$ wc -l $(find . -name "*.txt")
21022 ./writing/LittleWomen.txt
11 ./writing/haiku.txt
5 ./numbers.txt
21038 total
When the shell executes this command,
the first thing it does is run whatever is inside the $().
It then replaces the $() expression with that command’s output.
Since the output of find is the three filenames ./writing/LittleWomen.txt,
./writing/haiku.txt, and ./numbers.txt, the shell constructs the command:
$ wc -l ./writing/LittleWomen.txt ./writing/haiku.txt ./numbers.txt
which is what we wanted.
This expansion is exactly what the shell does when it expands wildcards like * and ?,
but lets us use any command we want as our own ‘wildcard’.
It’s very common to use find and grep together.
The first finds files that match a pattern;
the second looks for lines inside those files that match another pattern.
Here, for example, we can find txt files that contain the word “searching”
by looking for the string ‘searching’ in all the .txt files in the current directory:
$ grep "searching" $(find . -name "*.txt")
./writing/LittleWomen.txt:sitting on the top step, affected to be searching for her book, but was
./writing/haiku.txt:With searching comes loss
Matching and Subtracting
The
-voption togrepinverts pattern matching, so that only lines which do not match the pattern are printed. Given that, which of the following commands will find all .dat files increaturesexceptunicorn.dat? Once you have thought about your answer, you can test the commands in theshell-lesson-data/exercise-datadirectory.
find creatures -name "*.dat" | grep -v unicornfind creatures -name *.dat | grep -v unicorngrep -v "unicorn" $(find creatures -name "*.dat"))- None of the above.
Solution
Option 1. is correct. Putting the match expression in quotes prevents the shell expanding it, so it gets passed to the
findcommand.Option 2 is also works in this instance because the shell tries to expand
*.datbut there are no*.datfiles in the current directory, so the wildcard expression gets passed tofind. We first encountered this in episode 3.Option 3 is incorrect because it searches the contents of the files for lines which do not match ‘unicorn’, rather than searching the file names.
Binary Files
We have focused exclusively on finding patterns in text files. What if your data is stored as images, in databases, or in some other format?
A handful of tools extend
grepto handle a few non text formats. But a more generalizable approach is to convert the data to text, or extract the text-like elements from the data. On the one hand, it makes simple things easy to do. On the other hand, complex things are usually impossible. For example, it’s easy enough to write a program that will extract X and Y dimensions from image files forgrepto play with, but how would you write something to find values in a spreadsheet whose cells contained formulas?A last option is to recognize that the shell and text processing have their limits, and to use another programming language. When the time comes to do this, don’t be too hard on the shell: many modern programming languages have borrowed a lot of ideas from it, and imitation is also the sincerest form of praise.
The Unix shell is older than most of the people who use it. It has survived so long because it is one of the most productive programming environments ever created — maybe even the most productive. Its syntax may be cryptic, but people who have mastered it can experiment with different commands interactively, then use what they have learned to automate their work. Graphical user interfaces may be easier to use at first, but once learned, the productivity in the shell is unbeatable. And as Alfred North Whitehead wrote in 1911, ‘Civilization advances by extending the number of important operations which we can perform without thinking about them.’
findPipeline Reading Comprehension358242 Write a short explanatory comment for the following shell script:
wc -l $(find . -name "*.dat") | sort -nSolution
- Find all files with a
.datextension recursively from the current directory- Count the number of lines each of these files contains
- Sort the output from step 2. numerically
Key Points
findfinds files with specific properties that match patterns.
grepselects lines in files that match patterns.
--helpis an option supported by many bash commands, and programs that can be run from within Bash, to display more information on how to use these commands or programs.
man [command]displays the manual page for a given command.
$([command])inserts a command’s output in place.